Team:DTU-Denmark/Project testing sRNA
From 2011.igem.org
(→Strain construction) |
(→Strain construction) |
||
| Line 35: | Line 35: | ||
== Strain construction == | == Strain construction == | ||
| - | The strain used in testing our system is based on two strains: IG9 a ''E. coli'' based on the wild-type ''Escherichia coli'' K-12<span class="superscript">[[#References|[3]]]</span> strain W3110 with the ''lacZYA'', ''chbBCARG'' operons deleted, and IG302 which is the K-12 ''$ | + | The strain used in testing our system is based on two strains: IG9 a ''E. coli'' based on the wild-type ''Escherichia coli'' K-12<span class="superscript">[[#References|[3]]]</span> strain W3110 with the ''lacZYA'', ''chbBCARG'' operons deleted, and IG302 which is the K-12 ''$Delta$lacZYA'', ''$Delta$chiP'' (alias ''ybfM''), ''$Delta$chiX'' (alias ''sroB'', ''micM'') strain constructed by Rasmussen et al.<span class="superscript">[[#References|[12]]]</span>. |
Revision as of 20:19, 21 September 2011
Experiment: Testing sRNA
Contents |
Motivation
The biological experiments of the project is a proof of concept, showing that the E. coli sRNA sroB and the intergenic sRNA (trap-RNA) in the chbBCARG gene can be used to control gene expression by targeting the ybfM Shine-Dalgarno, and that these sRNAs can be rationally designed to target other Shine-Dalgarno sequences.
The experimental part consists of 3 distinct areas:
- Construction of plasmids
- Strain construction
- Improving the araBAD promoter
Construction of plasmids necessary for testing our system involves taking the native system from E. coli, as well as a slightly modified system and putting them into plasmids that let us both control the expression of these components and measure the output of the system.
Strain construction involves deleting the original genes from the chromosome of a E. coli W3110 strain. Since we use the original genes, these need to be deleted from the chromosome to prevent them from interfering with our measurements.
Improving the araBAD promoter entails expanding the dynamic range of this promoter by modifying the -10 and -35 sequence of the promoter, as well as randomly changing the nucleotide sequence around and in between these sequences. Since the araBAD promoter is used in our project improving this promoter could lead to even finer control of our system.
Construction of plasmids
In order to quantitatively describe the natural trap-RNA system the following three plasmids were constructed:
- PchiP-lacZ plasmid
- To mimic and test repression of the chiP gene its upstream region of 599 bp was fused to reporter genes lacZ and inserted on the pSB1C3 plasmid. Expression from PchiP promoter is costitutive however it can be induced to a higher level by chitobiose. LacZ codes for $\beta$-galactosidase and its activity was tested using $\beta$-Gal assay.
- Ptet-chiX plasmid
- ChiX, which is 84 bp long, was inserted into pSB4K plasmid together with regulatory element from tet operon. Expression from Ptet promoter was induced using anhydrotetracycline.
- PBAD-chiXR plasmid
- Introgenic region from chbBCARG operon 284 bp long was inserted into pSB3T5 plasmid. Expression from that plasmid was controlled by regulatory element from ara operon with arabinose as inducer.
Details on how the plasmids were constructed, including information on primers used, can be found in labnotebook.
Strain construction
The strain used in testing our system is based on two strains: IG9 a E. coli based on the wild-type Escherichia coli K-12[3] strain W3110 with the lacZYA, chbBCARG operons deleted, and IG302 which is the K-12 $Delta$lacZYA, $Delta$chiP (alias ybfM), $Delta$chiX (alias sroB, micM) strain constructed by Rasmussen et al.[12].
The three latter code for utilization of chitobiose (a sugar), chitobiose permease and the regulation of the chitobiose permease respectively.
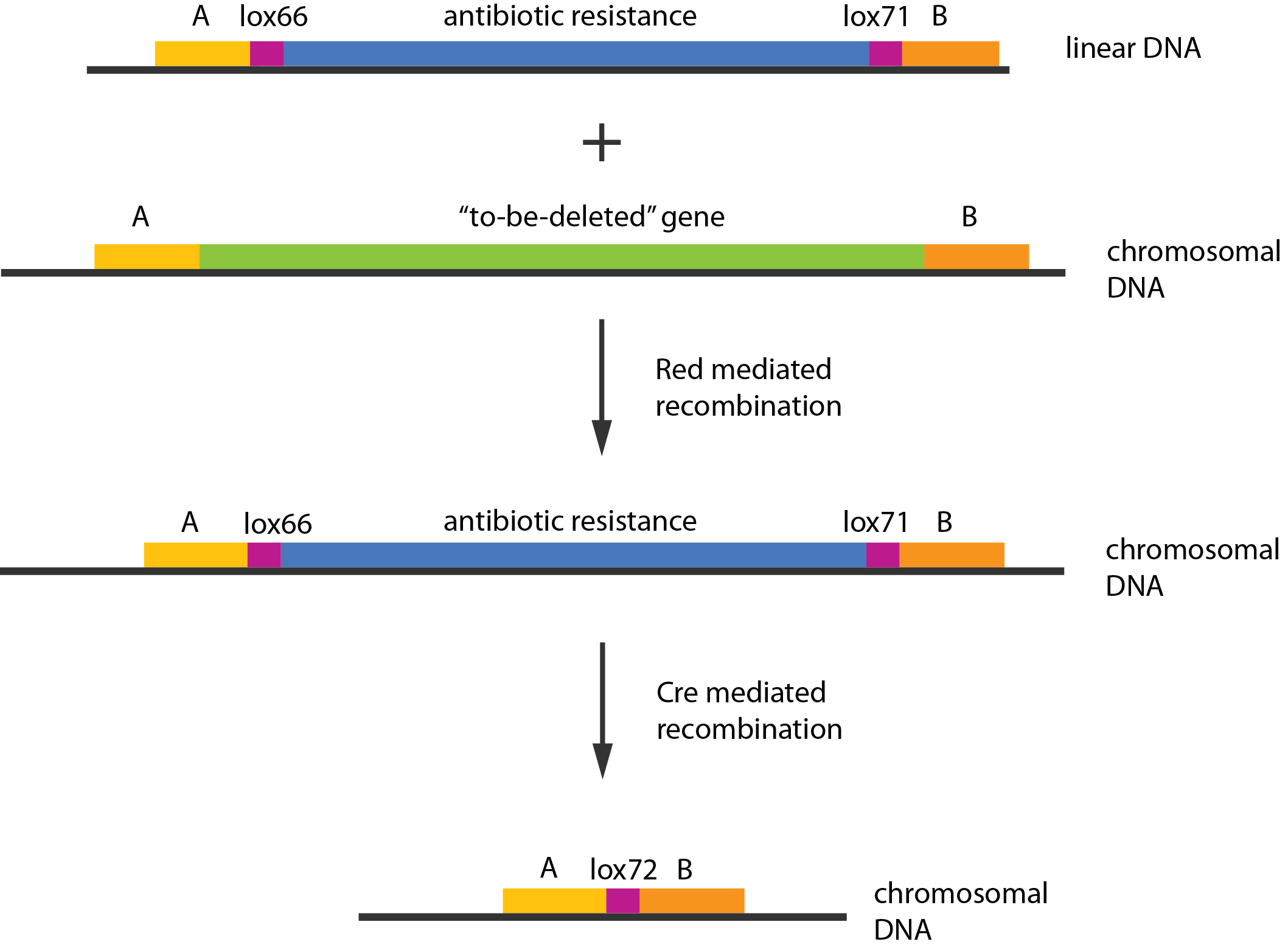
We used recombineering[1] to delete these genes. Schematically the procedure works in the following manner:
- Transform competent cells with Red and Cre helper plasmids.
- Make the cells competent and induce the Red recombination system.
- Transform with linear piece of DNA.
- Select for gain of resistance.
- Do colony PCR to check that resistance gene is inserted in the correct place.
- Move to medium inducing the Cre recombinase.
- Screen for loss of resistance.
- Verify the construct by colony PCR.
- Repeat step 2-7 as needed.
- Cure the helper plasmids.
The Red helper plasmids encodes the $\lambda{}$ phage Red recombination system. This system allows recombination events between sequences with around 40 nucleotides of homology. The cre helper plasmid encodes the Cre recombinase. This recombinase recognises 34bp loxP sequences and promotes recombination between them.
An example of a linear piece of DNA that could be used with this method, as well as the simplified representation of the procedure, can be seen in figure:
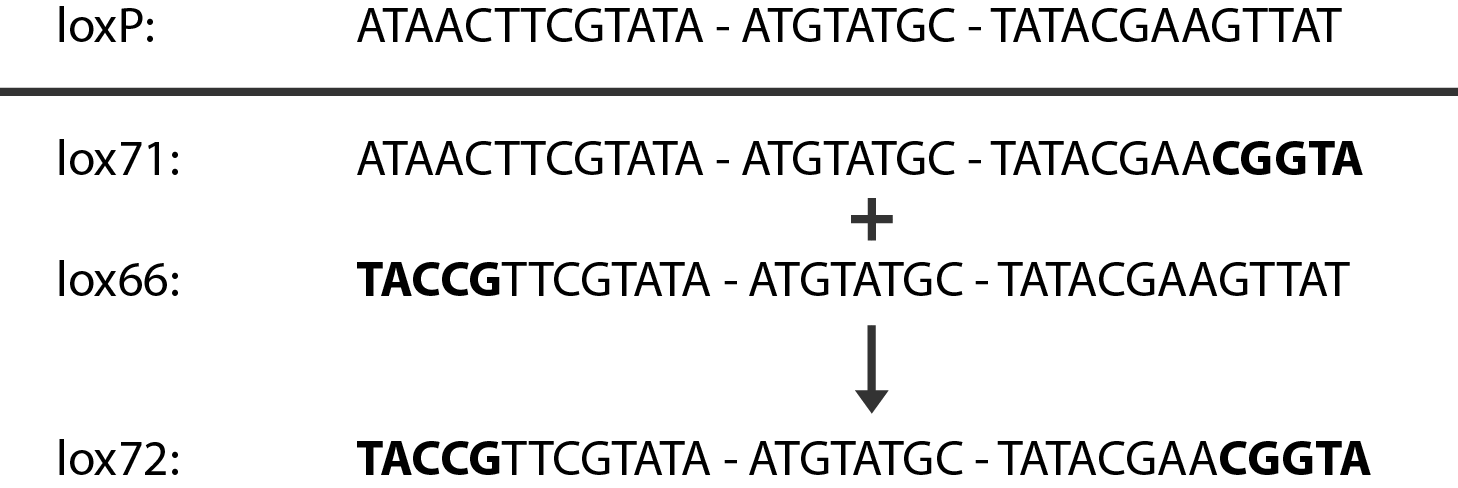
In case of two or more subsequent gene deletions using Red-Cre recombineering a problem of recombination between an old loxP site and newly introduced ones arises. Remember, that after each round of redswap one loxP site is left in the genome. As a result recombination using Cre recombinase in second or next redswap might not occur between loxP sites flanking a resistance gene. To circumvent this problem two mutated loxP sequences are used - lox66 and lox71[4]. In spite of the mutations these lox sites are recognized by Cre which can remove the intervening region. Finally, from lox66 and lox71 a new site is formed, lox72, which has a reduced affinity to Cre recombinase. Thus, more than one gene deletions can be subsequently conducted.
Improving the araBAD promoter
araBAD promoter
In our project to control sRNA chiXR expression we used the arabinose promoter (PBAD) from the araBAD operon that is regulated by araC protein[10]. AraC can work both as a repressor and activator of the PBAD and the mode of action depends on the presence of arabinose. PBAD promoter becomes activated in the presence of arabinose and repressed when arabinose is unavailable.
Choice of PBAD was not accidental. AraC repression in the absence of arabinose provides a very low level of background expression which can be lowered even more by adding glucose (catabolic repression). Once induced, this promoter allows a moderate level of expression. Finally, a range of arabinose concentrations can be used to achieve a desired level of induction[7][10].
There are however some known issues with PBAD promoter. Heterogeneous induction, known also as “all-or-nothing”[6] induction of cell culture is one of them. As a result only a subset of cells becomes induced with the arabinose. To circumvent “all-or-nothing” problem the arabinose transporter (AraE) could be placed under inducible promoter[7] or a different transporter could be engineered to have affinity to arabinose, e.g. LacY[11], and facilitates its uptake. Another problem arises when one wants to use PBAD promoter along with another promoter, e.g. PprpB promoter from propionate operon, and achieve a similar level of induction from both of them. This might not be feasible as PBAD is considerably weaker in expression than aforementioned PprpB promoter[8][10].
Approach
Since control of the system is essential to the applicability of our system we attempted to modify the dynamic range of the arabinose promoter ([http://partsregistry.org/Part:BBa_I0500 BBa_I0500]) by two means:
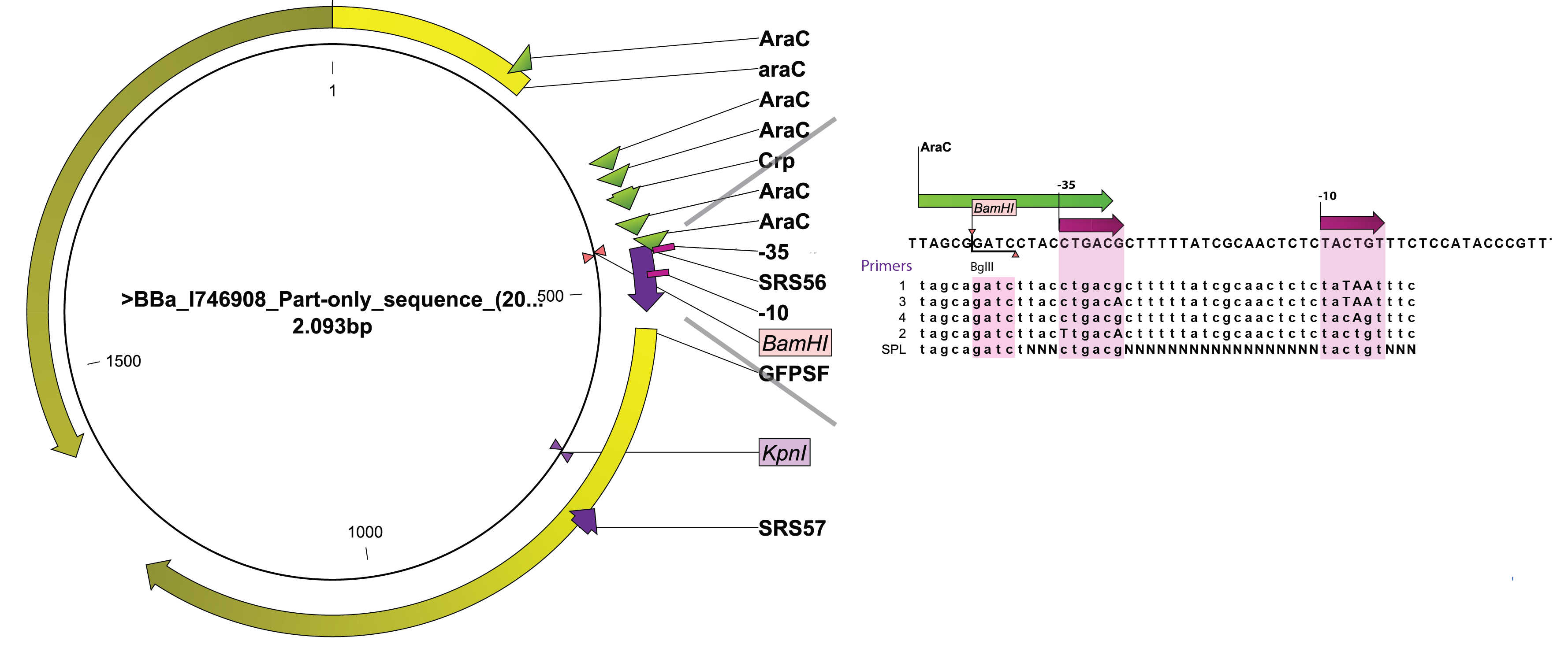
- rationally designing the -10 and -35 sequences of the promoter region since both the -35 (CTGACG) and the -10 (TACTGT) of the arabinose promoter are far from wild-type (TTGACA and TATAAT respectively).
- changing the nucleotides around and between the -10 and the -35 using synthetic promoter library (SPL), without changing the -10 and -35 from wildtype.
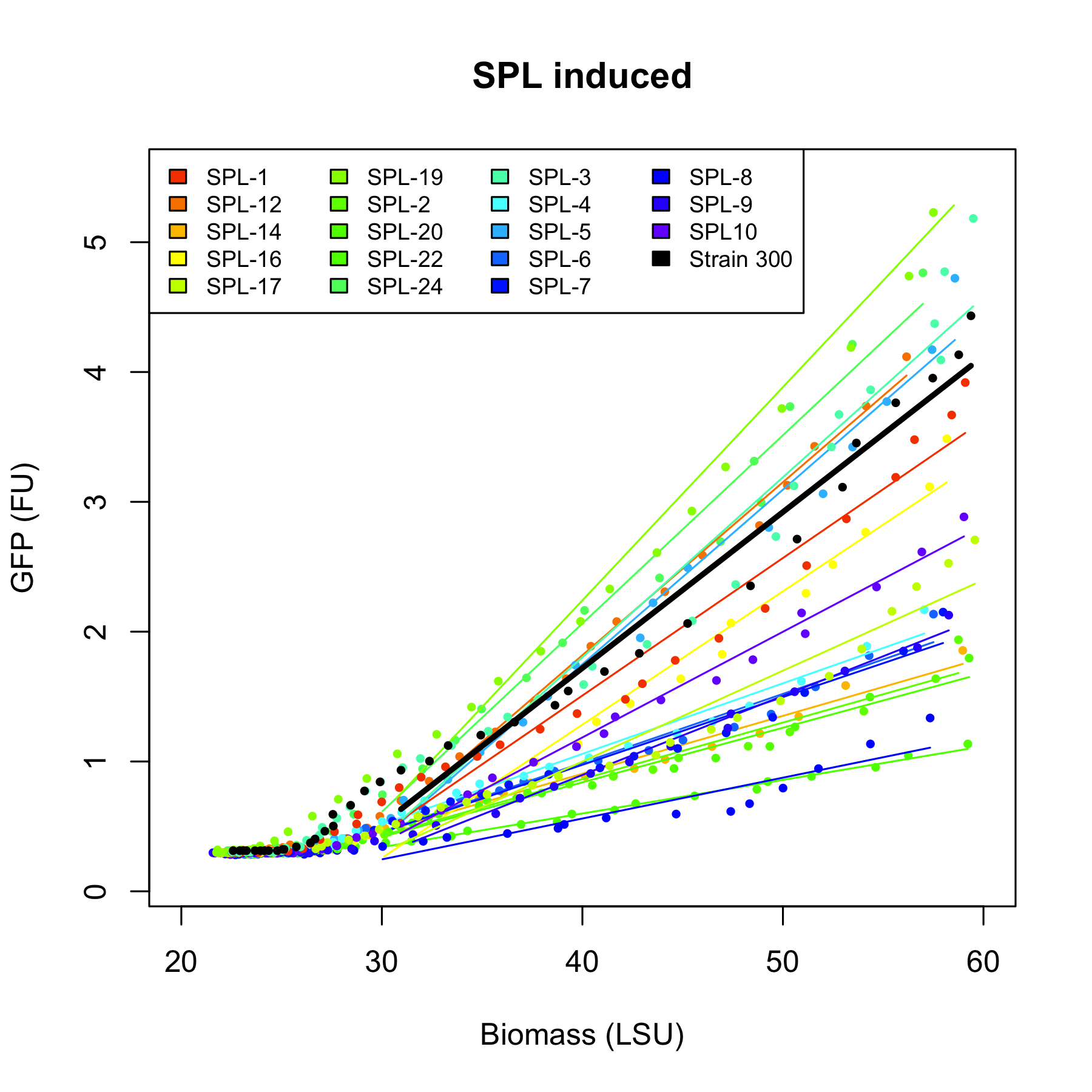
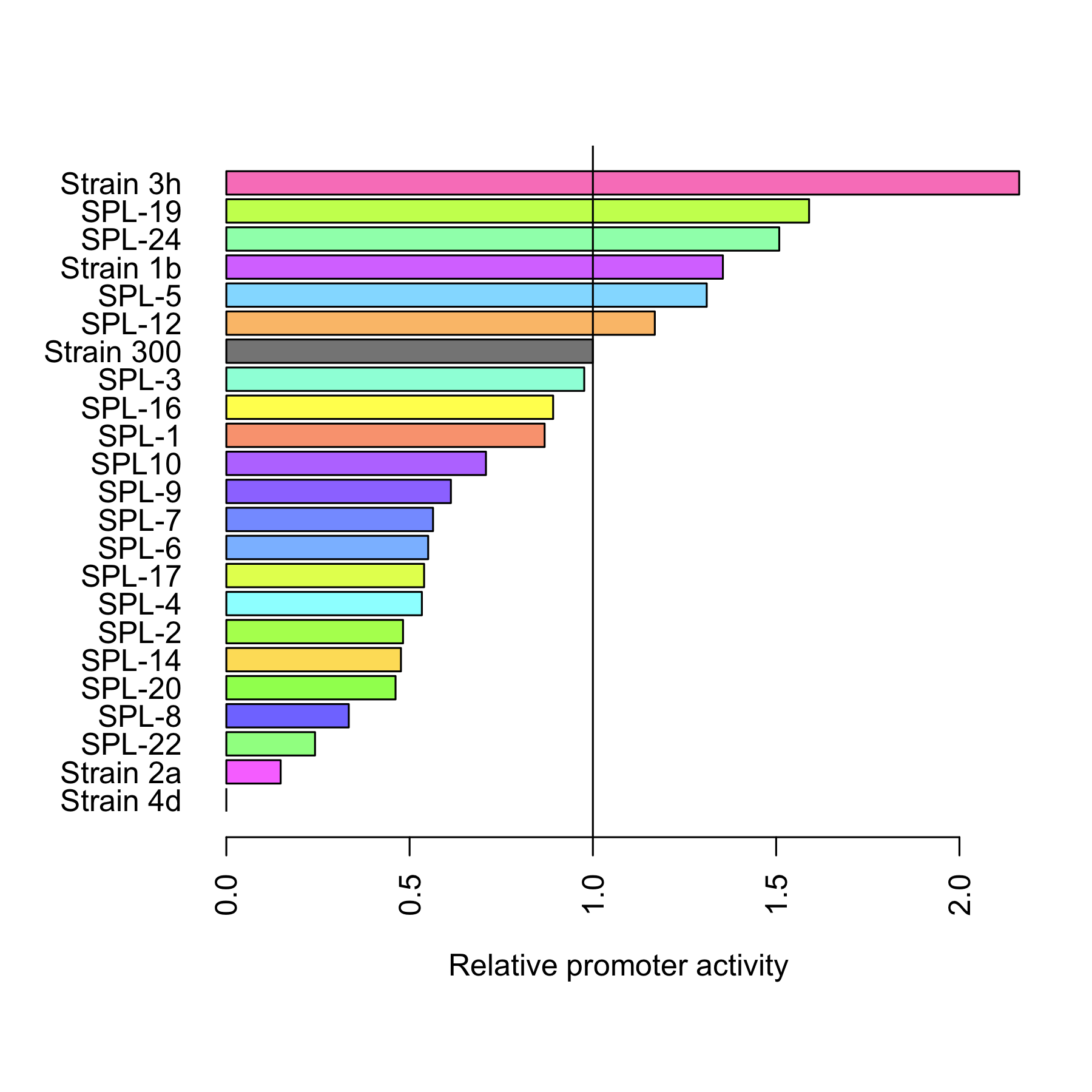
The arabinose promoter we were improving was fused to GFP and this construct came as a biobrick [http://partsregistry.org/Part:BBa_I746908 BBa_I746908]. To introduce the changes a set of primers was used to PCR PBAD. Four primers, SRS56 from 1 to 4, were used to make PCR products with mutated nucleotides within -10 and -35 and SRS56SPL primer was used to make the SPL PCR products. Then a round of restriction digestions and ligations followed which allowed to exchange the wild type PBAD with the mutated ones. Then they were transformed into E. coli strain DH5$\alpha$. Finally, all the rationally changed PBAD promoters and randomly selected SPL mutants were characterized using Biolektor.
Detailed description of the experiments, including information on primers used, can be found in labnotebook.
Results
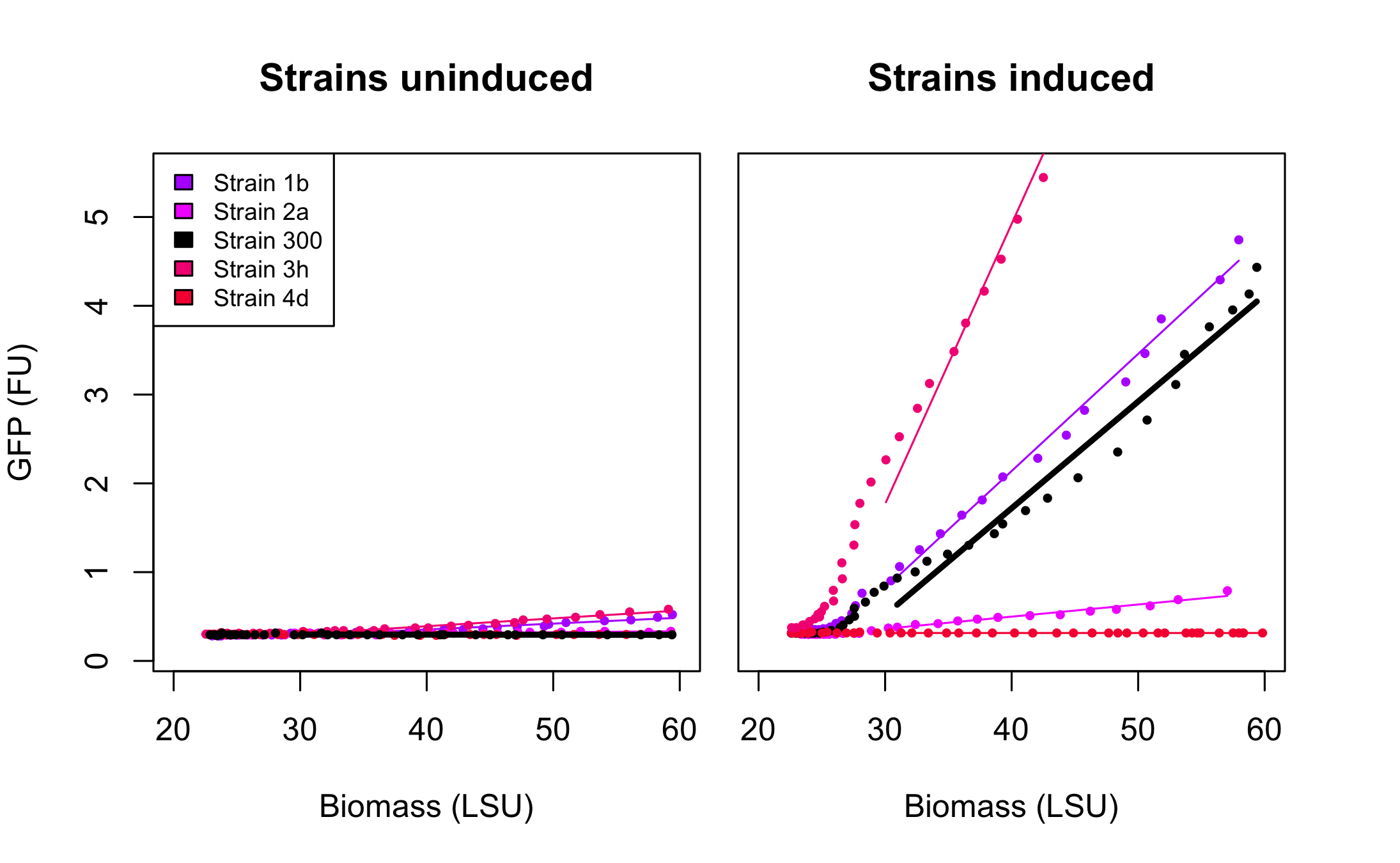
All 4 rationally designed (called Strain 1b, 2a, 3h, 4d) and 18 SPL-changed arabinose promoters (called SPL-1 up to SPL-24) fused to GFP were tested under induction of arabinose at concentration 0.2% and compared to non-induced state. Also, the original [http://partsregistry.org/Part:BBa_I746908 BBa_I746908] was characterized in the same fashion.
References
[1] Datta, Simanti, Nina Costantino, and Donald L Court. “A set of recombineering plasmids for gram-negative bacteria.” Gene 379 (September 1, 2006): 109-15. http://www.ncbi.nlm.nih.gov/pubmed/16750601.
[2] Figueroa-Bossi, Nara, Martina Valentini, Laurette Malleret, and Lionello Bossi. “Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target.” Genes & Development 23, no. 17 (2009): 2004 -2015.
[3] Hayashi, Koji, Naoki Morooka, Yoshihiro Yamamoto, Katsutoshi Fujita, Katsumi Isono, Sunju Choi, Eiichi Ohtsubo, et al. “Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110.” Molecular Systems Biology 2 (2006): 2006.0007. http://www.ncbi.nlm.nih.gov/pubmed/16738553.
[4] Lambert, Jolanda M, Roger S Bongers, and Michiel Kleerebezem. “Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum.” Applied and environmental microbiology 73, no. 4 (February 2007): 1126-35. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1828656&tool=pmcentrez&rendertype=abstract.
[5] Overgaard, Martin, Jesper Johansen, Jakob Møller‐Jensen, and Poul Valentin‐Hansen. “Switching off small RNA regulation with trap‐mRNA.” Molecular Microbiology 73, no. 5 (September 2009): 790-800.
[6] Novick, A. & Weiner, M. “Enzyme induction as an all-or-none phenomenon“ Proc. Natl. Acad. Sci. USA 43 (1957), 533–556.
[7] Khlebnikov, A., K. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. “Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter.” Microbiology (Reading, England) 147, no. Pt 12 (December 2001): 3241-7. http://www.ncbi.nlm.nih.gov/pubmed/11739756.
[8] Lee, Sung Kuk, and Jay D Keasling. “A Propionate-Inducible Expression System for Enteric Bacteria.” Society 71, no. 11 (2005): 6856-6862.
[9] Lee, Sung Kuk, and Jay D Keasling. “Propionate-Regulated High-Yield Protein Production in Escherichia coli.” Biotechnology (2005).
[10] Guzman, L M, D Belin, M J Carson, and J Beckwith. “Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter.” Journal of bacteriology 177, no. 14 (July 1995): 4121-30. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=177145&tool=pmcentrez&rendertype=abstract.
[11] Morgan-Kiss, Rachael M, Caryn Wadler, and John E Cronan. “Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity.” Proceedings of the National Academy of Sciences of the United States of America 99, no. 11 (May 28, 2002): 7373-7. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=124238&tool=pmcentrez&rendertype=abstract.
 "
"