Team:DTU-Denmark/Project
From 2011.igem.org
| Line 16: | Line 16: | ||
'''Construction of plasmids''' necessary for testing our system involves taking the native system from ''E. coli'', as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system. | '''Construction of plasmids''' necessary for testing our system involves taking the native system from ''E. coli'', as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system. | ||
| + | |||
'''Strain construction''' involves deleting the original genes from the chromosome of a \textit{E. coli} W3110 strain. Since we use the original genes, these need to be deleted from the chromosome to prevent them from interfering with our measurements. | '''Strain construction''' involves deleting the original genes from the chromosome of a \textit{E. coli} W3110 strain. Since we use the original genes, these need to be deleted from the chromosome to prevent them from interfering with our measurements. | ||
| + | |||
'''Improving the ''araBAD'' promoter''' entails expanding the dynamic range of this promoter by modifying the -10 and -35 sequence of the promoter, as well as randomly changing the nucleotide sequence around and in between these sequences. Since the ''araBAD'' promoter is used in our project improving this promoter could lead to even finer control of our system. | '''Improving the ''araBAD'' promoter''' entails expanding the dynamic range of this promoter by modifying the -10 and -35 sequence of the promoter, as well as randomly changing the nucleotide sequence around and in between these sequences. Since the ''araBAD'' promoter is used in our project improving this promoter could lead to even finer control of our system. | ||
{{:Team:DTU-Denmark/Templates/Standard_page_end}} | {{:Team:DTU-Denmark/Templates/Standard_page_end}} | ||
Revision as of 15:53, 18 September 2011
Project
Tuning regulation with a non-coding RNA trap
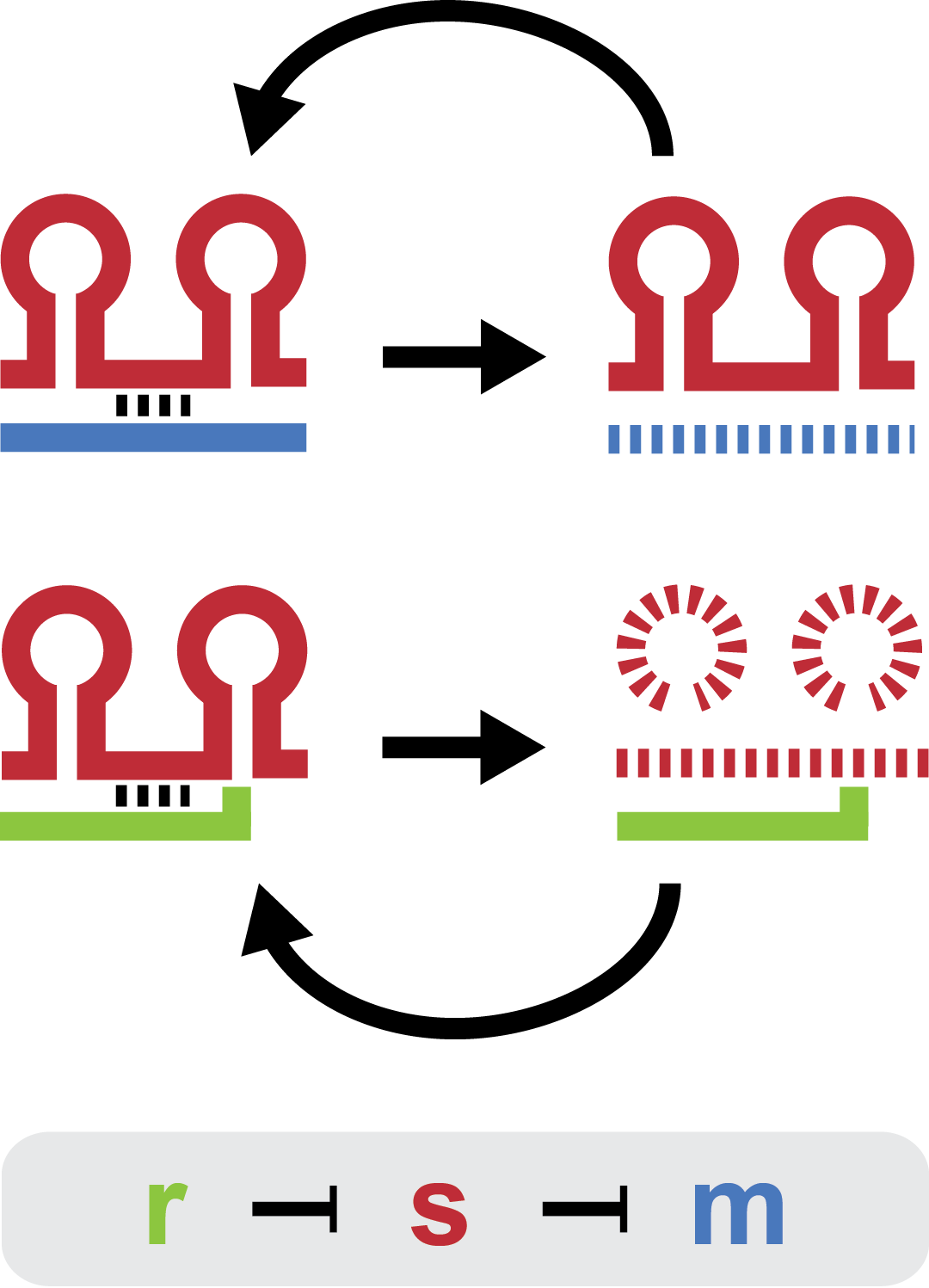
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. One such application could be the optimization and fine-tuning of synthetic biological circuits, which is currently a cumbersome process of trial and error. We have investigated a novel type of RNA regulation, where the inhibition caused by a small regulatory RNA is relieved by another RNA called trap-RNA. The system displays a large dynamic range and can uniquely target and repress any gene of interest providing unprecedented flexibility. We suspect that any level of repression is achievable by simply altering the sequences of the involved RNAs. Multiple such systems can coexist without interfering and are thus compatible with more complex designs. Furthermore the trap-RNA can be fused to any transcript in effect allowing any gene to act as an activator.
Overview
Our project is a proof of concept project, showing that the E. coli sRNA sroB and the intergenic sRNA in the chbBCARG gene can be used to control gene expression by targeting the ybfM Shine-Delgarno, and that these sRNAs can be rationally designed to target other Shine-Delgarnos.
The experimental part of the project can be broken into 3 distinct parts, which combined form the complete project:
- Construction of plasmids
- Strain construction
- Improving the araBAD promoter
Construction of plasmids necessary for testing our system involves taking the native system from E. coli, as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system.
Strain construction involves deleting the original genes from the chromosome of a \textit{E. coli} W3110 strain. Since we use the original genes, these need to be deleted from the chromosome to prevent them from interfering with our measurements.
Improving the araBAD promoter entails expanding the dynamic range of this promoter by modifying the -10 and -35 sequence of the promoter, as well as randomly changing the nucleotide sequence around and in between these sequences. Since the araBAD promoter is used in our project improving this promoter could lead to even finer control of our system.
 "
"