Team:DTU-Denmark/Background
From 2011.igem.org
| (83 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:DTU-Denmark/Templates/Standard_page_begin| | + | {{:Team:DTU-Denmark/Templates/Standard_page_begin|Overview}} |
| - | = | + | <div class="overviewPage"> |
| - | + | ||
| - | + | <div class="overviewBox left"> | |
| + | == Small RNA == | ||
| + | [[File:DTU1_Central_dogma_with_regulators.png|200px|right]] | ||
| + | In our project we dig into the fascinating world of small RNA (sRNA) regulation with the aim of engeneering a so-called trap sRNA regulating module and hereby creating a universal tool for a whole new way of regulating gene expression in synthetic biology. To fully understand our project, at basic knowledge of sRNAs, the way they regulate gene expression, the players involved (the RNA chaperone Hfq) and the unique features of the regulation is required. So continue reading if these words are new to you. | ||
| - | + | [https://2011.igem.org/Team:DTU-Denmark/Background_sRNA Read more...] | |
| + | </div> | ||
| + | <div class="overviewBox right"> | ||
| - | + | == The chitobiose system == | |
| + | [[File:Chitobiose_system.png|200px|right]] | ||
| - | + | The project is inspired by the regulation of chitobiose uptake and metabolism in E. coli. Especially the following three players, which have been indentified independently in several organisms and thus goes by many names... | |
| + | : 1. The '''mRNA''' for a chitoporin that transports chitosugars into cells, called '''''chiP''''' (alias ''ybfM'') | ||
| + | : 2. A '''sRNA''' which regulates the chitoporin post-transcriptionally, called '''''chiX''''' (alias ''sroB'', ''micM'') | ||
| + | : 3. A '''trap-sRNA''' which is transcribed from intergenic region in the chitobiose operon | ||
| + | : (chbBCARG), which we named '''''chiXR''''' for ''chiX'' regulator. | ||
| + | : [https://2011.igem.org/Team:DTU-Denmark/Background_the_natural_system Read more...] | ||
| + | |||
| + | </div> | ||
| + | <div class="overviewBox left"> | ||
| + | |||
| + | == Synthetic biology == | ||
| + | [[File:DTU_HumanInteractome1.png|200px|right]] | ||
| + | Synthetic biology is a scientific field concerning the design of new biological functions. The way humans do design is fundamentally different than the evolutionary principles from which life origins. Life has evolved due to an incredible long series of trial-and-error driven and governed by selectional pressure. As this approach have led to an enormous variety of solutions to ways of being alive on earth, from the most sophisticated machine known to mankind, the human brain, to the most resilient microorganisms, there is no doubt to the success of evolution. | ||
| + | |||
| + | [https://2011.igem.org/Team:DTU-Denmark/Background_synthetic_biology Read more...] | ||
| + | </div> | ||
| + | |||
| + | <div style="clear:both;"></div> | ||
| + | </div> | ||
{{:Team:DTU-Denmark/Templates/Standard_page_end}} | {{:Team:DTU-Denmark/Templates/Standard_page_end}} | ||
Latest revision as of 03:14, 22 September 2011
Overview
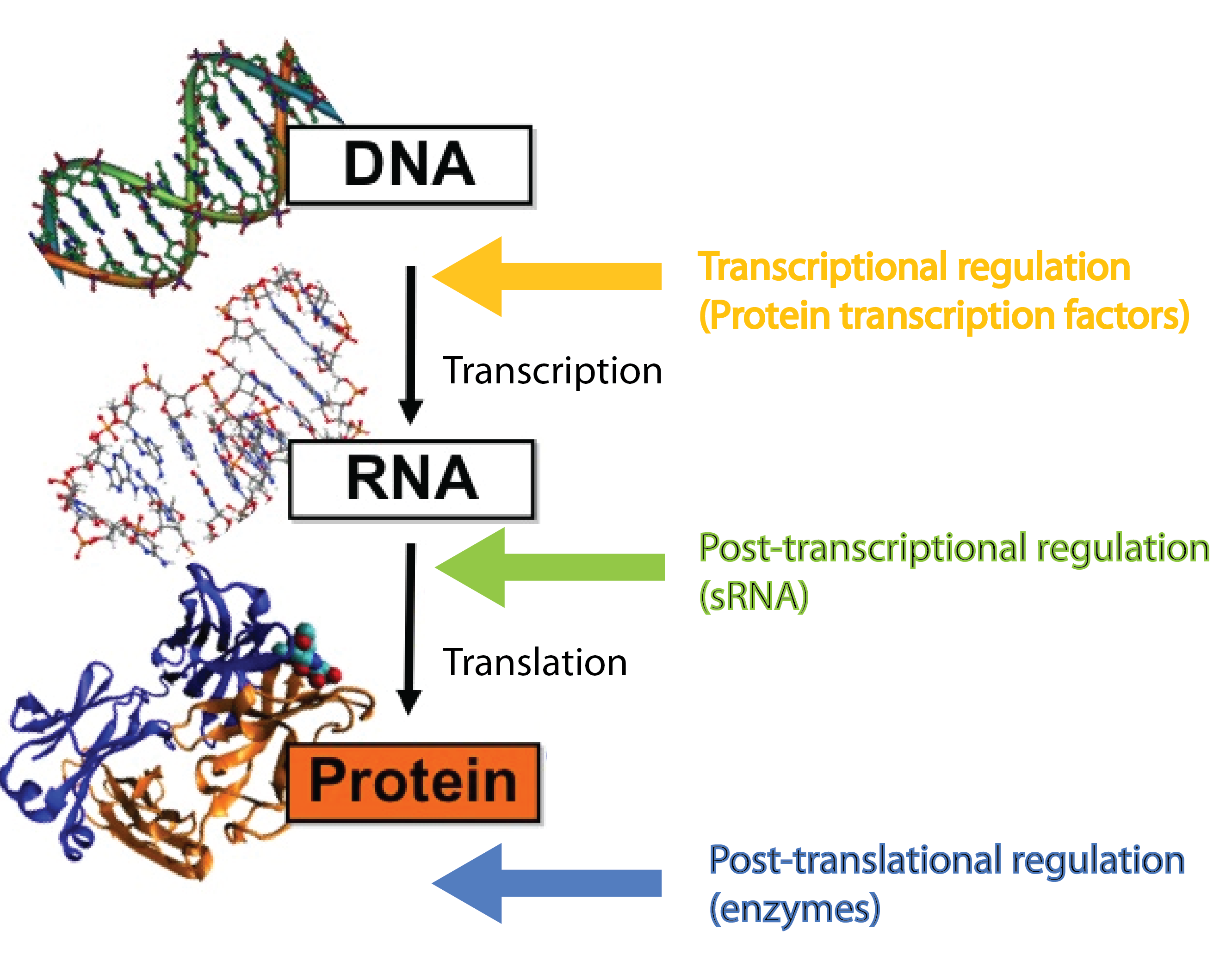
Small RNA
In our project we dig into the fascinating world of small RNA (sRNA) regulation with the aim of engeneering a so-called trap sRNA regulating module and hereby creating a universal tool for a whole new way of regulating gene expression in synthetic biology. To fully understand our project, at basic knowledge of sRNAs, the way they regulate gene expression, the players involved (the RNA chaperone Hfq) and the unique features of the regulation is required. So continue reading if these words are new to you.
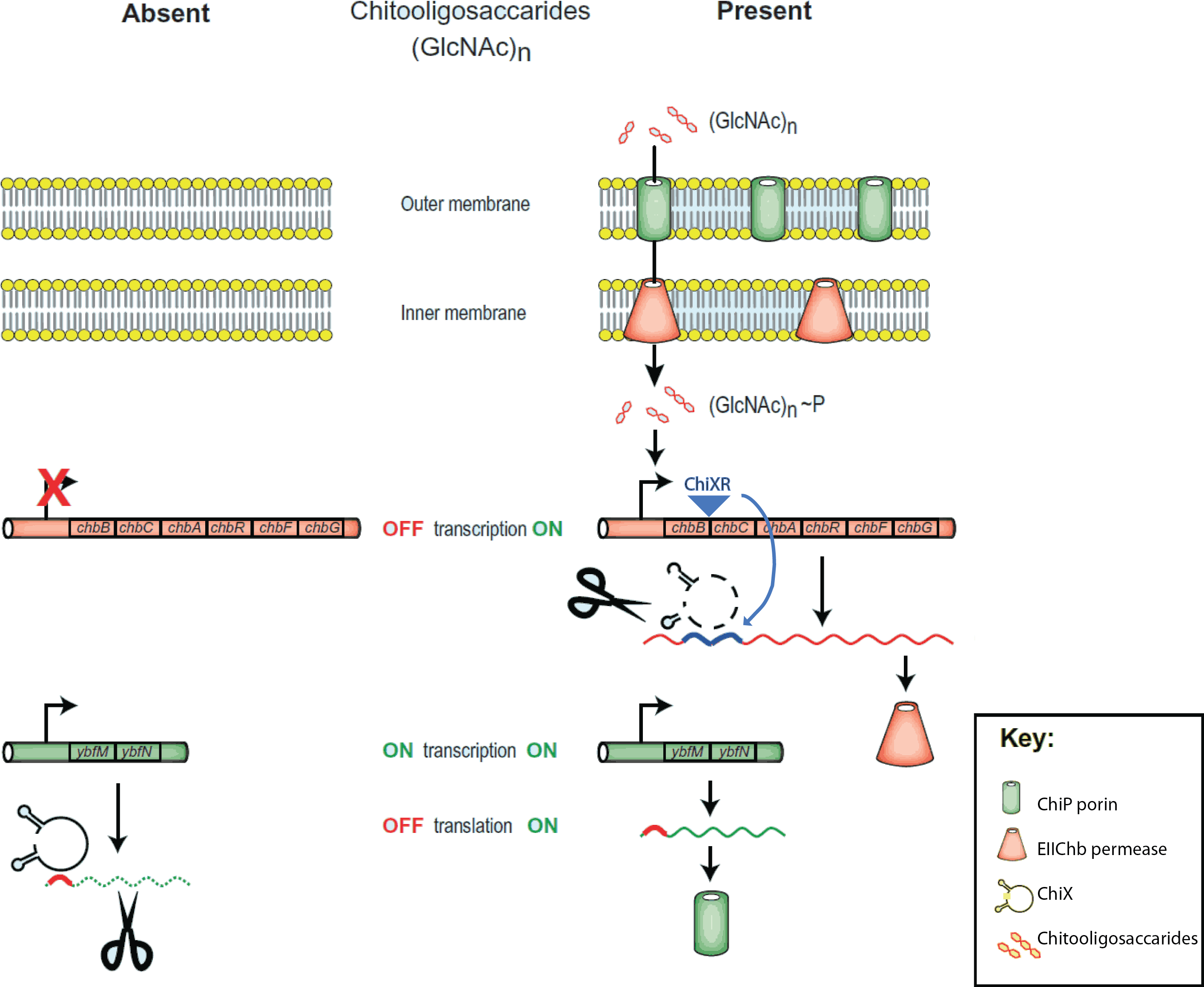
The chitobiose system
The project is inspired by the regulation of chitobiose uptake and metabolism in E. coli. Especially the following three players, which have been indentified independently in several organisms and thus goes by many names...
- 1. The mRNA for a chitoporin that transports chitosugars into cells, called chiP (alias ybfM)
- 2. A sRNA which regulates the chitoporin post-transcriptionally, called chiX (alias sroB, micM)
- 3. A trap-sRNA which is transcribed from intergenic region in the chitobiose operon
- (chbBCARG), which we named chiXR for chiX regulator.
- Read more...
Synthetic biology
Synthetic biology is a scientific field concerning the design of new biological functions. The way humans do design is fundamentally different than the evolutionary principles from which life origins. Life has evolved due to an incredible long series of trial-and-error driven and governed by selectional pressure. As this approach have led to an enormous variety of solutions to ways of being alive on earth, from the most sophisticated machine known to mankind, the human brain, to the most resilient microorganisms, there is no doubt to the success of evolution.
 "
"