Team:Bielefeld-Germany/Labjournal
From 2011.igem.org
(Difference between revisions)
(→Week 1: 2nd - 8th may) |
(→Week 12: 18th july - 24th july) |
||
| Line 127: | Line 127: | ||
| - | Molecular Beacons: | + | [[https://2011.igem.org/Team:Bielefeld-Germany#NAD.2B_detection | Molecular Beacons]]: |
* cloning the NAD+ dependent ligase from ''E. coli'' into BioBrick backbones | * cloning the NAD+ dependent ligase from ''E. coli'' into BioBrick backbones | ||
Revision as of 12:27, 9 August 2011
On this page we summarize the (successful) results and achievements of our teamwork.
Week 1: 2nd - 8th may
- cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic)
- cloning of fusionprotein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter
- testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection
- expression of the successfully assembled BPA degrading BioBricks in E. coli [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10]
- successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html Brevibacterium flavum] and Corynebacterium crenatum
- successful cloning of the S-layer gene of B. flavum without TAT-sequence
Organizational:
- meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld
Week 2: 9th - 15th may
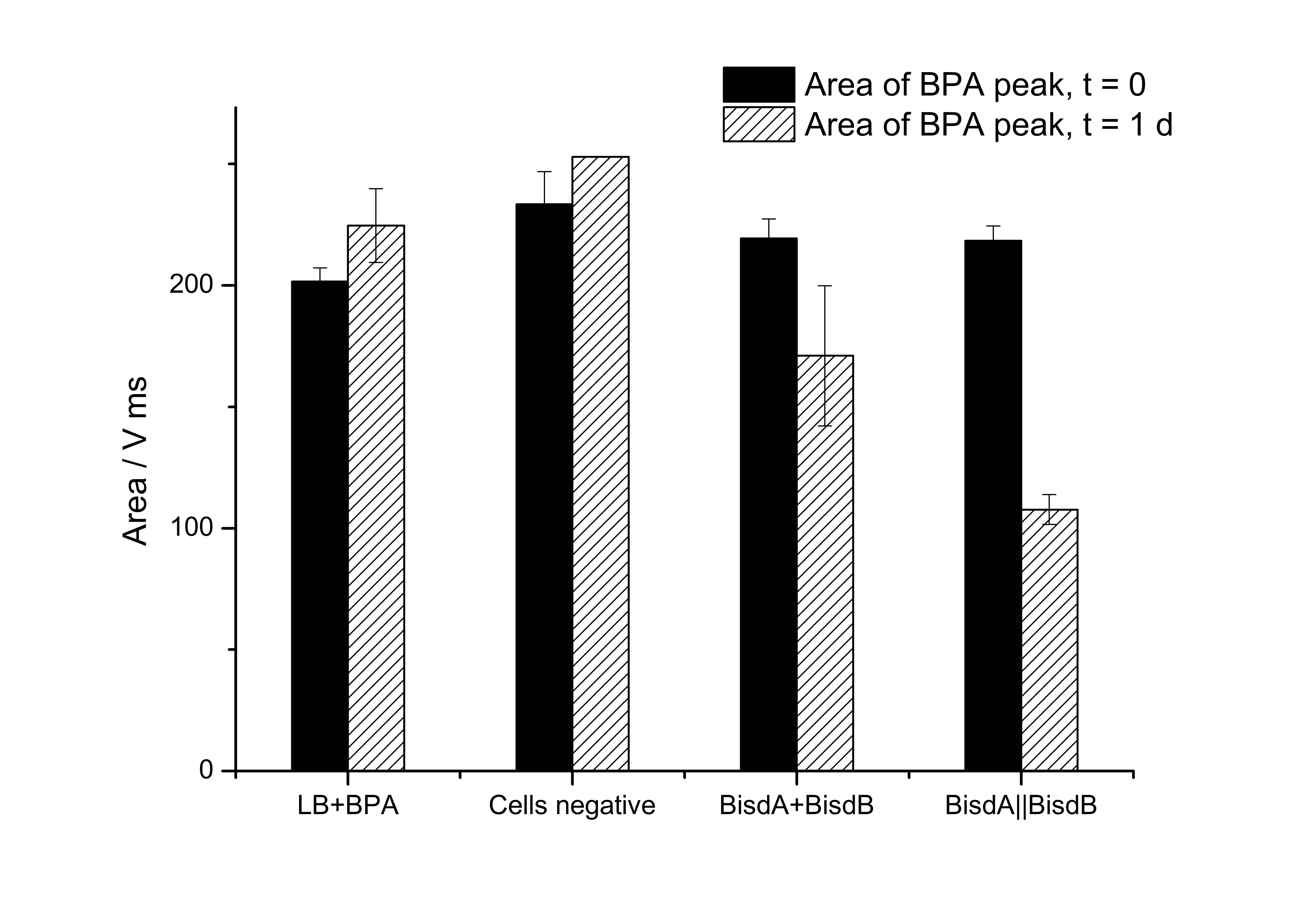
Figure 1: HPLC results of the first experiment on BPA degradation in E. coli TOP10. Cultivations were carried out in LB medium with 100 mg L-1 BPA at 30 °C. Samples were taken at the beginning of the cultivation and after one day. The HPLC results are shown above (area of the BPA peak). BisdA + BisdB is the polycistronic gene and BisdABisdB is the fusion protein.
Bisphenol A:
- assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system)
- HPLC results: fusionprotein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare fig. 1)
S-layer:
- expression of the S-layer gene without TAT-sequence of B. flavum in E. coli [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results
- successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 Corynebacterium halotolerans]
Organizational:
- moving to our own room in the [http://www.cebitec.uni-bielefeld.de/ CeBiTec]
Week 3: 16th - 22nd may
Bisphenol A:
- successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP oxidoreductase] gene from E. coli TOP10
S-layer:
- successful cloning of the complete S-layer gene cspB of B. flavum, C. crenatum and C. halotolerans
- successful cloning of the S-layer genes of B. flavum and C. halotolerans without TAT-sequence, without lipid anchor and without both (only self-assembly domain)
Organizational:
Week 4: 23rd - 29th may
Bisphenol A:
- establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS)
S-layer:
Organizational:
- arrange a BBQ for our workgroup in the CeBiTec to get to know our coworkers
- substantiating our contribution to the GENIALE
Week 5: 30th may - 5th june
Bisphenol A:
- beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>)
S-layer:
Organizational:
- meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in july
Week 6: 6th - 12th june
Organizational:
- presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention]
Week 7: 13th - 19th june
Bisphenol A:
- cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites
Week 8: 20th - 26th june
Organizational:
- finishing our first press release
- all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived
Week 9: 27th june - 3rd july
S-Layer:
- our synthesized S-Layers SgsE and SbpA finaly arrived
Week 10: 4th july - 10th july
Bisphenol A:
- analyzing the influence of temperature, promotor intensity and the characteristics of the fusion protein on BPA degradation
Organizational:
- presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research).
Week 11: 11th july - 17th july
Bisphenol A / S-Layer:
- our BioBrick order (some fluorescences proteins and cleavage sites) from iGEM HQ arrived
Organizational:
- presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos])
Week 12: 18th july - 24th july
Organizational:
- presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld
- meeting and having fun with the iGEM teams from Delft/NL, Edinburgh/UK, Odense/DK. Freiburg/DE and Ljubljana/SL at the Symposium ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627300144576/ Photos])
Bisphenol A / S-Layer:
- removing illegal restiction sites to get valid BioBricks
- cloning the NAD+ dependent ligase from E. coli into BioBrick backbones
Week 13: 25th july - 31th july
S-Layer:
- fusing the synthesized S-Layers to a bunch of fluorescent proteins
Bisphenol A:
- testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F)
Week 14: 1st august - 7th august
Bisphenol A:
- BPA analysis with extraction and LC-MS finally works and is very accurate
- Better results for BPA degradation in E. coli -> our fusion protein (<partinfo>K123000</partinfo> to <partinfo>K123001</partinfo>) can completely degrade BPA
- Measuring characterization results for different BPA degrading BioBricks
Molecular Beacons:
- Successful characterization of two differently labeled Molecular Beacons as a preparation for the NAD+ bioassay including autonomously produced NAD+ dependent DNA ligase from E. coli
Week 14: 8th august - 14th august
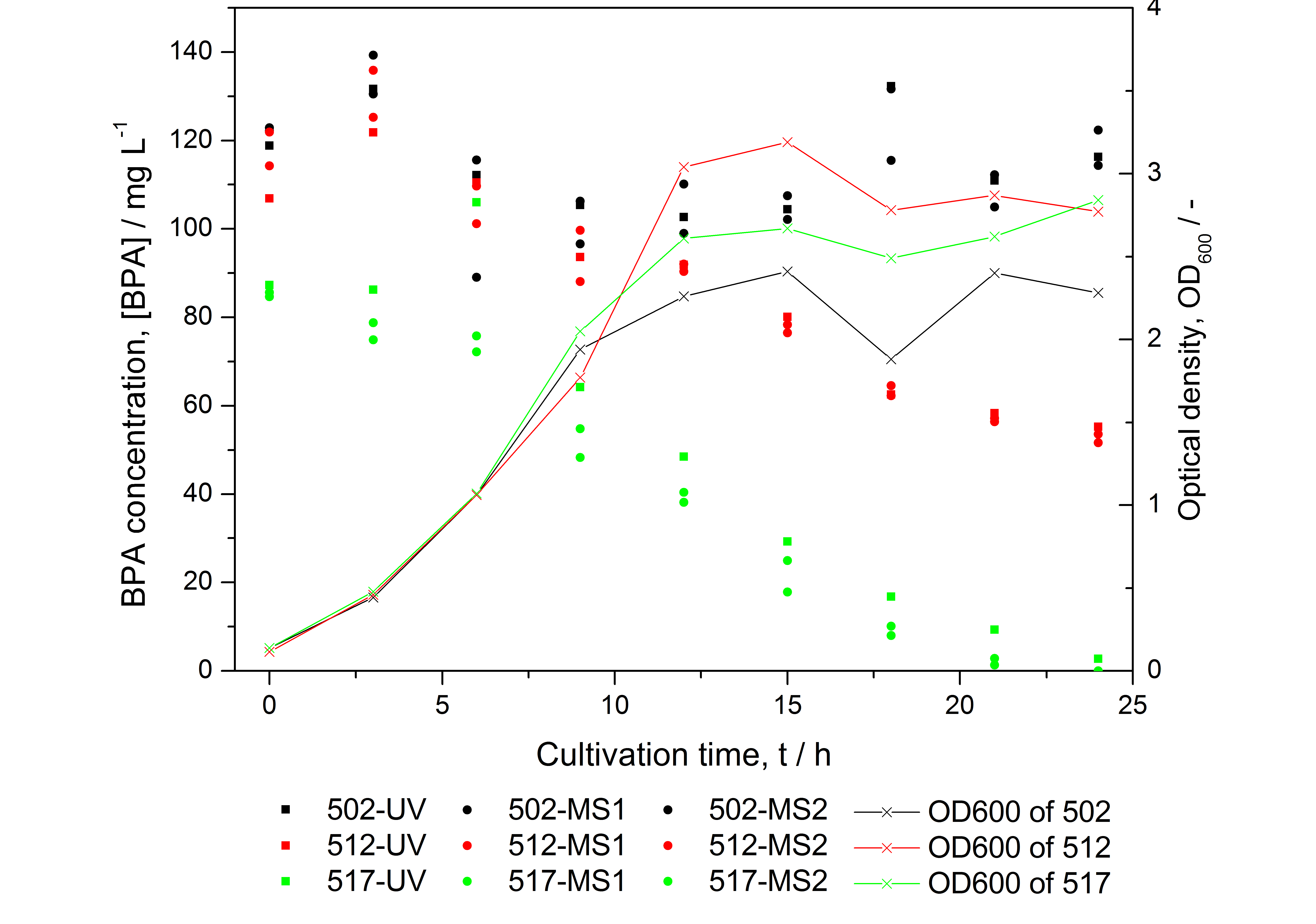
Figure 1: HPLC results and optical density of experiments on BPA degradation of E. coli KRX. Cultivations were carried out in LB medium with ~ 100 mg L-1 BPA at 30 °C. Samples were taken every three hours over one day. BPA concentration was measured by HPLC with either UV detector or ESI-qTOF-MS. 502: negative control (<partinfo>K123000</partinfo>), 512: polycistronic <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, 517: fusionprotein between <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, every part behind medium strong constitutive promoter.
Bisphenol A:
- BPA analysis with extraction and HPLC with UV detector leads to the same results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is higher than of "normal" HPLC, compare figure to the right)
- measuring of more samples from cultivations with BPA degrading BioBricks for further characterization
 "
"