Yeast Vector Library
Overview
Due to Saccharomyces cerevisiae's amenability to genetic manipulation as well other benefits associated with a eukaryotic host, there has recently been an increase in interest regarding yeast as a microbial workhorse. With this comes a need for synthetic biological tools to manipulate yeast quickly and easily. The goal of this sub-project is to create a library of vectors that both efficiently manipulates DNA in S. cerevisiae as well as conforms to the BioBrick Standard Assembly protocol. In designing these vectors, we hope to utilize the speed of propagation of E. coli along with the production capabilities of S. cerevisiae to create the first easy-to-use BioBrick-standard vector series.
pRS Series Vectors
The library will be a slight modification of the pRS series vectors created by Sikorski et al.[1] and Christianson et al.[2]. They are designed to be "shuttle vectors", which can be easily propagated in Escherichia coli as well as transfected into the final chassis, S. cerevisiae. They are a hybrid of a yeast vector and the E. coli phagemid vector pBluescript SK+, which contains a lacZ gene with a polylinker as well as an ampicillin resistance (bla) gene. They contain an E. coli origin of replication (the pBluescript SK+ ori) and for the episomal plasmids, a yeast origin (either CEN/ARS or 2µ). Sikorski et al. and Christianson et al. designed three types of vectors: Integrating, CEN/ARS and 2μ (2micron) vectors:
- Integrating Plasmids: A combination of the YIp (Yeast Integrating Plasmid) type vector with the multipurpose E. coli pBluescript II SK+ phagemid. Chromosomally integrates the plasmid.
- CEN/ARS Plasmids: Endogenous yeast plasmids based on the YCp (Yeast Centromere plasmid) type plasmid as well as the previously mentioned pBluescript II SK+. Medium copy number plasmid.
- 2µ Plasmids: YEp (Yeast Episomal plasmid) type plasmid. On the backbone of pBluescript II SK+ phagemid. High copy number plasmid of about 20 per cell.
Within each vector type, individual vectors differ by four selectable markers genes: HIS3 (essential for histidine biosynthesis), TRP1 (tryptophan), LEU2 (leucine), and URA3 (uracil). Each individual vector (12 total) requires a specific mutant strain of yeast with the respective biosynthesis gene knocked out. For E. coli colony selection, all the vectors contain LacZ genes for the white/blue ß-galactosidase assay.
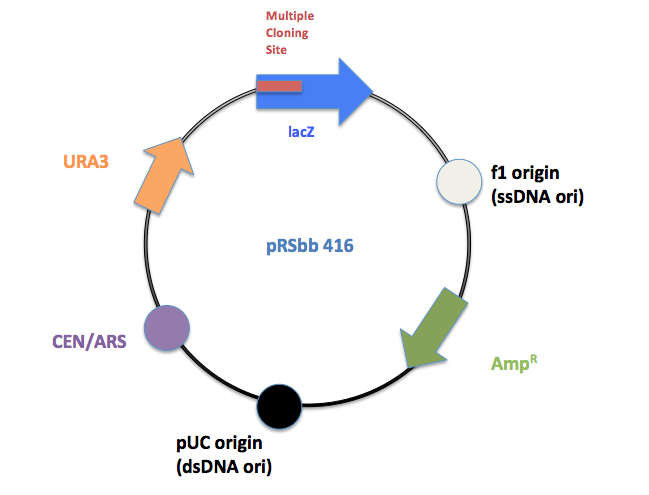
pRSbb Series Vectors
The new BioBrick Standard vectors (pRSbb) contain the necessary restriction enzymes in the correct order in the place of the original pRS-series multiple cloning site. These vectors will be ready to accept any part via the BioBrick Standard Assembly protocol:
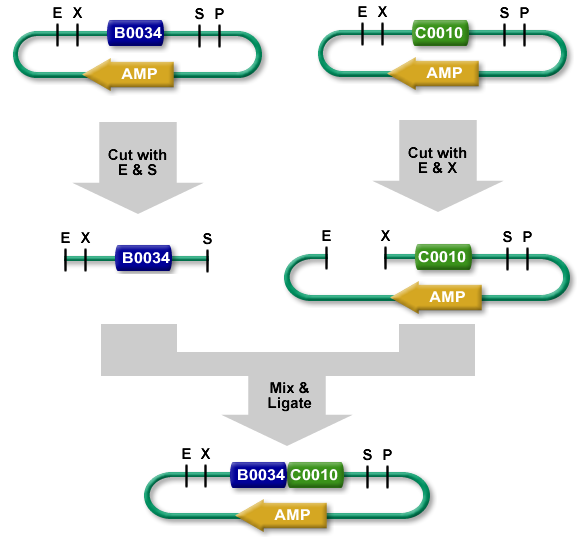
BioBrick Standard Assembly. E = EcoRI, X = XbaI, S = SpeI, P = PstI
Final Vectors
| pRSbb # | Type | Marker type |
| 403 | Integrating | HIS3 |
| 404 | Integrating | TRP1 |
| 405 | Integrating | LEU2 |
| 406 | Integrating | URA3 |
| 413 | CEN/ARS | HIS3 |
| 414 | CEN/ARS | TRP1 |
| 415 | CEN/ARS | LEU2 |
| 416 | CEN/ARS | URA3 |
| 423 | 2µ | HIS3 |
| 424 | 2µ | TRP1 |
| 425 | 2µ | LEU2 |
| 426 | 2µ | URA3 |
Workflow
Removal of restriction enzyme sites from vector
Any instances of restriction enzymes used in the BioBrick protocol must be removed from the vector.
- Find the occurrence of EcoRI, XbaI, PstI, and SpeI sites in all of the pRS400-series vectors.
- Create a spreadsheet with these values, compare them to Dr. Boeke’s numbers
- Map out all of the vectors including ORFs, RE sites and origins of replication and annotate their respective sequences in ApE files
- Comb through all of these annotated sequences and find one vector for each type of plasmid with the least number of changes necessary.
- Criteria for removal of RE sites (Quick Change):
- RE site is in ORF, ori, or other impt. Feature
- Set up QuikChange protocol for the 3 initial vectors
- Notes:
- Look at translation map to determine which two (or three) codons the RE sites translates to
- Change the 3rd “wobble base” of a codon
- Make a conservative change to a codon of similar bias to the changed codon or most used codon.
- Notes:
- Finalize list of changes. To include:
- What to change
- Where change occurs
- Change to what
- How to change (primer info, etc.)
- Order primers
- Perform [http://kirschner.med.harvard.edu/files/protocols/Stratagene_quickchangepdf.pd QuikChange® PCR]
- Record and store all data on a backup
- Gel verification
- Transform into E. coli
- Make minipreps; cut with enzyme of choice
Conversion from existing to BioBrick compatible multiple cloning site
- Determine correct prefix/suffix
- Design primers to remove MCS and replace with BioBrick Standard
- Order primers
- PCR
- Gel verification
- Transform
- Miniprep and cut
- Send winners from above to sequencing
- Make Perms (freeze cells in 15% glycrol, stored at -80C
- Conclude, Clean up, Write report and Aggregate Data
Primer Design
Multiple cloning site removal and insertion
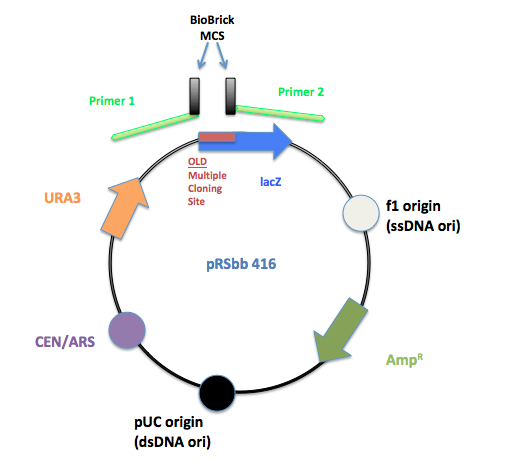
PCR primers were designed to flank the regions of the old pRS multiple cloning site and replace it with the BioBrick prefix and suffix.
The initial pRS 416 with primers flanking the multiple cloning site to remove the old MCS and insert the new BioBrick MCS.
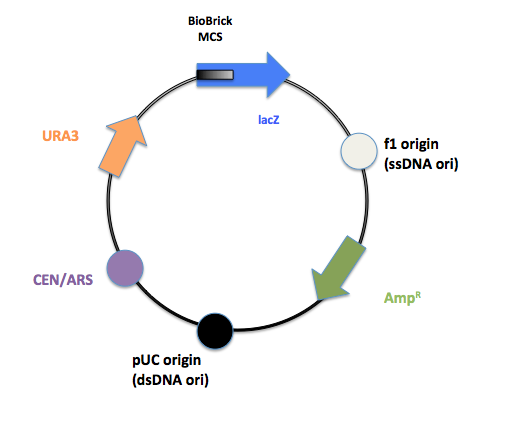
The final product after PCR.
Site directed mutagenesis
In order to ensure the vector will be cut in only the correct places by specific restriction enzymes, instances of certain restriction enzyme sequences must be removed from the vector. The restriction enzymes required for BioBrick Standard Assembly are:
- EcoRI: GAATTC
- XbaI: TCTAGA
- SpeI: ACTAGT
- PstI: CTGCAG
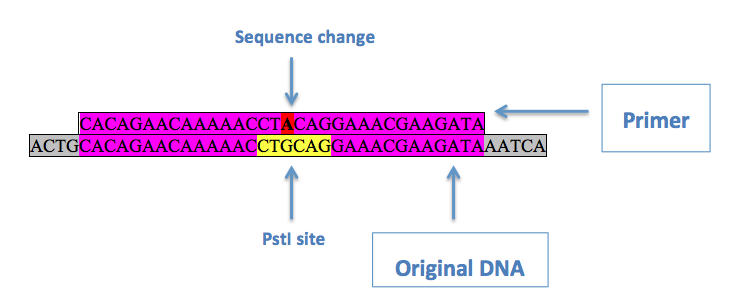
If a restriction enzyme sequence that needed to be changed was in an open reading frame, the third ("wobble") base of an in-frame codon within the sequence was changed to ensure a silent mutation. This is done using site directed mutagenesis. This involves a primer with the desired mutation in the middle, with regions of homology on either side:
Above is an example of a primer to remove a PstI site from pRS416. Purple denotes regions of homology, red is the desired base change, and yellow is the original restriction enzyme site. Two primers are used, one being the reverse complement of the other, with the [http://kirschner.med.harvard.edu/files/protocols/Stratagene_quickchangepdf.pdf QuikChange® Site-Directed Mutagenesis] protocol in order to substitute the desired base in.
References
[1]Sikorski, R.S.; Hieter, P. (1989) [http://www.ncbi.nlm.nih.gov/pubmed/2659436 "A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae.".] Genetics. 1989 May; 122(1):19-27.
[2] Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. (1992) [http://www.ncbi.nlm.nih.gov/pubmed/1544568 "Multifunctional yeast high-copy-number shuttle vectors".] Gene. 1992 Jan 2; 110(1):119-22.
 "
"