Team:Edinburgh/Phage Display
From 2011.igem.org
Phage Display: Proposals
The second system our feasibility study will examine, while searching for a way to keep extracellular enzymes close together, is based on displaying proteins on a bacteriophage. This type of display is called "phage display".
Contents |
Outline
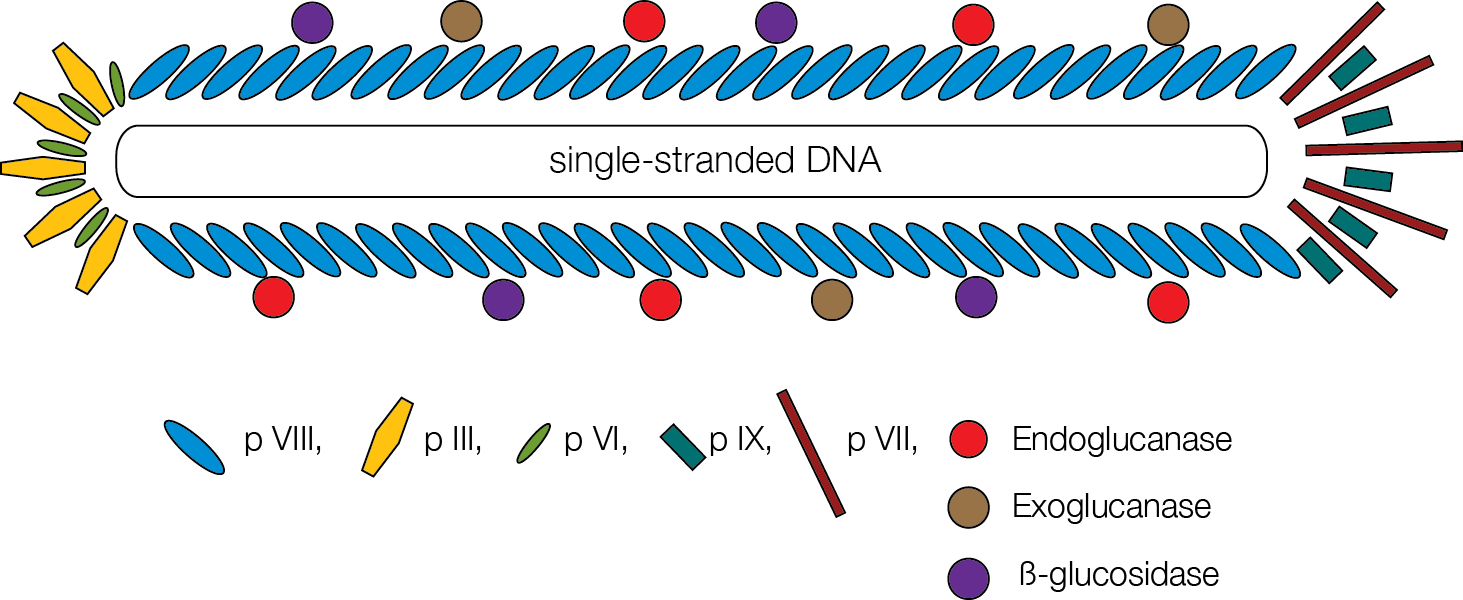
The project will use phage M13, which is non-lytic (does not kill the bacteria). The entire genome is known (e.g. [http://genome-www.stanford.edu/vectordb/vector_descrip/COMPLETE/M13MP18.SEQ.html here] is the entire M13mp18 genome) so primers can be designed. New England Bioworks [http://www.neb.com/nebecomm/tech_reference/protein_tools/phdFaq.asp#1.6 claims]:
- "The major coat protein pVIII is present at ~2700 copies per virion, of which ~10% can be reliably fused to peptides or proteins."
We would infect our E. coli with wildtype phage. Our E. coli would have a plasmid coding for a fusion between an enzyme and pVIII. Proteins can be fused to pVIII at its amino terminal (i.e. 5' end in the DNA), according to [http://www.utoronto.ca/sidhulab/pdf/08.pdf Weiss and Sidhu, 2000]. pVIII has a leader peptide ([http://www.uniprot.org/uniprot/P69541 residues 1-23]) that is cleaved out, slightly complicating fusion design.
To attach several different proteins to the phage, different fusions can be created and all of them expressed on the plasmid.
We would need to tune expression levels of the fusions versus the wildtype protein.
The F plasmid
M13 can only infect E. coli if it has a sex pilus. This is coded for by the F plasmid. However, such plasmids are quickly lost from cultures in a laboratory setting, as they confer a fitness cost. The JM109 strain has proline synthesis genes (proAB) absent from its main chromosome, but present on the F plasmid, and so when it is grown on minimal media, the plasmid will not be lost, as it is needed to keep the bacteria alive.
Genetic instability
In order to display several different proteins on one phage, it will be necessary to have several copies of the pVIII gene fused to different enzymes. The presence of repeated sequences on a plasmid can lead to genetic instability.
This will not be a problem in the JM109 lab strain, which lacks an important recombination enzyme. As for the use of this technology in industry, it will be possible to overcome this problem simply by synthesising coding sequences with as many altered (but synonymous) codons as possible. We have written a software tool for designing such sequences... see the genetic instability page.
Bead reactors
Maurice Gallagher, one of our university's phage experts, suggests a different approach: make several strains of bacteria each producing phage with just one type of pVIII-fusion. But also make each phage have a pIII-fusion with a protein which could bind some sort of bead. The bead now becomes the complete "reactor". Since each bacteria codes for only one pVIII fusion, there is no repeated sequence problem.
Problems
The question is how efficiently fusions to pVIII can get onto the phage. There are some dire warnings in the literature:
- In our experience, most large proteins display well below one copy per phage particle. — [http://www.sciencedirect.com/science/article/pii/S0022283699934654 Sidhu et al (2000)]
- A large 20 kDa protein (human growth hormone, hGH) is not displayed at detectable levels. — [http://www.utoronto.ca/sidhulab/pdf/08.pdf Weiss and Sidhu (2000)]
- The properties of the pIV channel may be one of the factors that limit the size of polypeptides that can be displayed on pVIII. — [http://www.immun.lth.se/fileadmin/immun/Avhandlingar/Fredrik_Karlsson.pdf Karlsson (2004)]
- As a general rule, the minor coat proteins will display larger proteins more effectively than pVIII. — [http://pubs.acs.org/doi/full/10.1021/cr000261r Kehoe and Kay (2005)]
- [It is plausible] that a phage containing pVIII with a large peptide may be too large in diameter to pass through the 7-nm pIV exit pore in the outer membrane. — Barbas et al (2001)
- The pVIII site, although very popular for peptide phage display, is not suitable for the efficient display of large polypeptides such as antibodies. — [http://www.sciencedirect.com/science/article/pii/S0734975000000549 Benhar (2001)]
However, Maurice seemed quite upbeat about the prospects when we met him. It's definitely the case that large stuff has been displayed, but the question is whether this is some sort of fluke that only occurs (say) once per phage (i.e. see the Sidhu quote above).
A possible solution: zipper adaptors
If things don't work well, a possible solution (or at least something to attempt) exists...
MIT 2010 attached small "zipper" peptides to pVIII. Their intention was to join phage together. We could use this system to attach our enzymes of interest (i.e. our cellulases) to the phage indirectly, by fusing the enzyme to a zipper and pVIII to the corresponding zipper. The two zippers will attach if we can get them to meet physically.
The basic idea is similar to that of [http://www.sciencedirect.com/science/article/pii/S0378111905000764 Paschke and Höhne (2005)] (see Figure 1) and especially [http://www.sciencedirect.com/science/article/pii/S0022283609014624 Wang et al (2010)].
- Sequence for zipper GR1: EEKSRLLEKENRELEKIIAEKEERVSELRHQLQSVGGC (38)
- Sequence for zipper GR2: TSRLEGLQSENHRLRMKITELDKDLEEVTMQLQDVGGC (38)
The construct would then contain two coding sequences:
- Promoter--RBS--Leader--GR2--pVIII
- Promoter--RBS--Periplasm Signal--Enzyme--GR1
Testing
As proof of concept (i.e. something we can accomplish in a short time) it would be sufficient to get just one fusion protein working. We need to prove that the enzyme part is actually getting out of the cell, so we must demonstrate that some substance which cannot enter the cell is nevertheless being degraded.
A fairly easy test would be to use a fusion of amylase to pVIII, and assay for starch degradation. There is no amylase BioBrick (with DNA available) in the Registry, so we'd have to make it.
Example systems
A simple version of the system would work as follows:
- E. coli are grown up containing a plasmid coding for a pVIII fusion gene, i.e:
- Promoter--RBS--Leader--Amylase--(Linker?)--pVIII.
- These E. coli are infected with M13.
- They create new phage; some of the modified pVIII proteins incorporate into the capsid.
More complex versions would either incorporate more pVIII fusions, or multiple strains all of which have a pIII-fusion to attach to beads.
Conceptual animation
Results
Please see the team's Data Page for information about how far we got with this project.
References
- Barbas CF, et al (2001) Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press.
- Benhar I (2001) [http://www.sciencedirect.com/science/article/pii/S0734975000000549 Biotechnological applications of phage and cell display]. Biotechnology Advances 19(1): 1-33 (doi: 10.1016/S0734-9750(00)00054-9).
- Paschke M, Höhne W (2005) [http://www.sciencedirect.com/science/article/pii/S0378111905000764 A twin-arginine translocation (Tat)-mediated phage display system]. Gene 350(1): 79-88 (doi: 10.1016/j.gene.2005.02.005).
- Sidhu SS, Weiss GA, Wells JA (2000) [http://www.sciencedirect.com/science/article/pii/S0022283699934654 High copy display of large proteins on phage for functional selections]. Journal of Molecular Biology 296(2): 487-495 (doi: 10.1006/jmbi.1999.3465).
- Wang KC, Wang X, Zhong P, Luo PP (2010) [http://www.sciencedirect.com/science/article/pii/S0022283609014624 Adapter-directed display: a modular design for shuttling display on phage surfaces]. Journal of Molecular Biology 395(5): 1088-1101 (doi: 10.1016/j.jmb.2009.11.068).
- Weiss GA, Sidhu SS (2000) [http://www.utoronto.ca/sidhulab/pdf/08.pdf Design and evolution of artificial M13 coat proteins]. Journal of Molecular Biology 300: 213-219 (doi: 10.1006/jmbi.2000.3845).
 "
"