Team:Harvard/Results/Chip Synthesis
From 2011.igem.org
qPCR curves
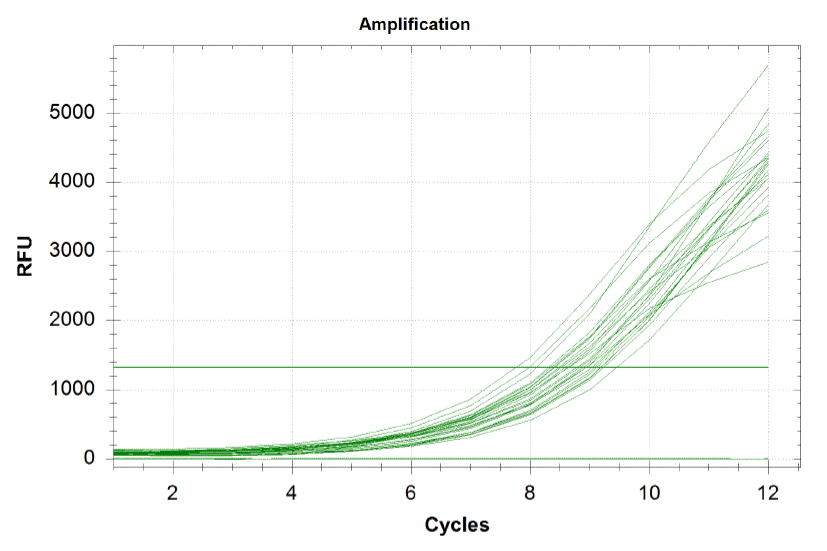
When amplifying the oligos, one wants to perform a qPCR in order to monitor the growth rates of each sub-pool and ensure that growth is happening at equal rates. By plotting the relative flouresence present in each cycle, we can visualize the pogress of the reaction. One wants to run the reaction while the growth is still exponential. During this phase, all the oligos are replicated at equal rates. Once growth begins leveling off, as it does at the end of the graph below, the reaction should be stopped as the oligos are now being replicated unequally. Stopping the reaction at the very end of the exponential growth phase ensures that we do not have significantly more oligos of a particular sequence in our pool, and decreases the bias in our results.
Each line on the graph below represents a qPCR for one of our sub-pools (each sequence we are targeting represents a sub-pool: CB top, CB bottom, FH top, FH bottom, Myc 198, Myc 981).
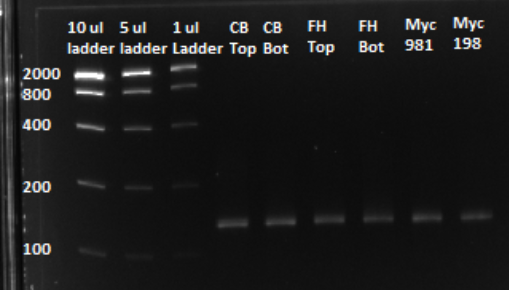
Based on the graph and gel below, we knew we successfully amplified each of our target sub-pools.
Because flourescence grows exponentially and just begins to level off at the end of the graph (where PCR was terminated), we are confident that overamplification did not occur, so individual oligos in each of our sub-pools were amplified at roughly equal rates. This preserves library integrity
After a PCR clean up, we ran a gel in order to confirm that the qPCR produced the expected product, of approximately 130 bps. Based on the gel, we could conclude that our qPCR was indeed successful, and our oligo library was ready to use.
Sequencing results of Library transformation
Error rates
A common question that rises when dealing with novel synthesis techniques concerns the error rates. After all, if chip synthesis produced oligos in which errors were ubiquitous, then the technique would be of little practical value, despite the low cost. In order to understand what the error rates of our chip were, we sent out a 96 plate of post-transformation colonies for sequencing. Of those, 77 had sequencing results. We then compared the sequences of these oligos to the Finger 1 oligos that were originally ordered.
As seen above 44 out of 77, or 57.1%, of our sequences were perfect matches.
This means there were two problems we then had to deal with: point mutations and frameshift mutations.
In total, 20.8% of our oligos contained point mutations. However, point mutations in the helix region are unlikely to affect the structure of the the protein. In fact, they add additional variation to our F1 library. It is entirely possible that one of these mutants ends up being a strong binder and allows the cell to survive selection, whereas the original sequence is not. It should also be noted that the actual rate of point mutations is lower than those documented. We simply looked as the nucleotide sequence of the oligos, not the trace files. It is possible that some reported point mutations have bad traces and thus, may not actually represent a SNP.
The 17 frame shift mutations (22.1%) are a bigger problem, since that means we have essentially lost an oligo we wanted to test. It is possible there are other copies of the oligo without the error, and that our cell happened to uptake one with a detrimental mutation. In any case, cells that take up plasmids that contain an F1 region with a frameshift will not survive selection.
We determined the overall per base pair error rate for this set sequenced to be around 1/200, which includes errors generated by the chip, or generated during PCR and assembly. These are a bit higher than those found by Kosuri, et al., but within a reasonable margin.
Distributions
Of the 77 samples with good sequencing results, 2 sequences were repeated once. Discounting these, 73 of the 77 sequences, or 94.8%, were unique. This suggests that there is substantial variability within the library, and then we should not be concerned that our library is dominated primarily by a single oligo.
 "
"