Team:Macquarie Australia/Characterisation
From 2011.igem.org
Contents |
Characterisation of our Biobrick
The Phytochromes
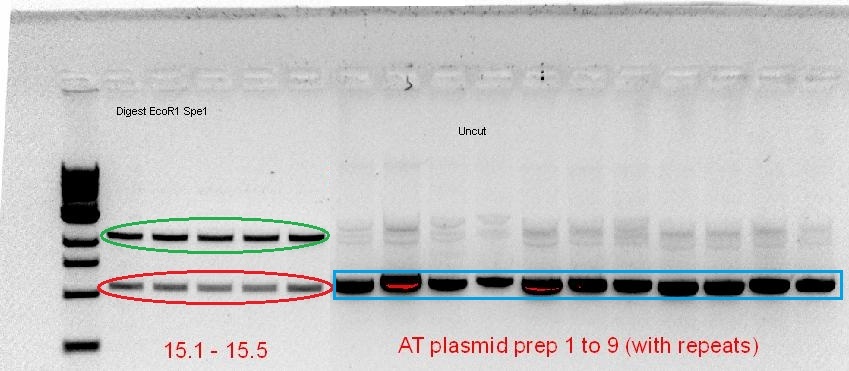
The starting template that we had to work with was a RBS-coupled phytochrome coding sequence containing plasmid (for A. tumefaciens) and PCR product (for D. radiodurans) from the 2010 Macquarie iGEM team. Using the BioBrick compatible primers designed, PCR optimisation for the 2 phytochromes was screened over a range of temperatures, buffer types and cycle conditions (see notebook). When the desired product size was visible on the gel, the band was cut out and gel purified, subsequently following the BioBrick construction pipeline as mentioned above. However, after the screening of the purified plasmid from the transformed cells, it seemed that the plasmid contained either no insert (for AT-Bph) or the wrong insert (for DR-Bph).
Green - plasmid backbone, Red - Size of ~1kb, corresponds with size of Red fluorescent protein, Blue - Size of plasmid show that there is no insert (based on previous experience, see notebook)
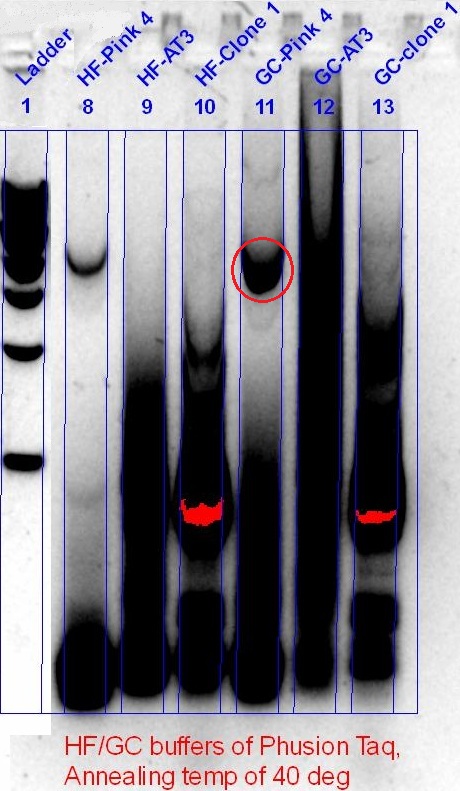
Despite having alkaline phosphatase treatment, and gel purification of the cut plasmid backbone (not shown), the re-ligation of the backbone, and the appearance of the red fluorescent protein, showed that the BioBrick construction of the phytochromes failed. Hence, PCR optimisation to get a better product was performed, this time with a much lower annealing temperature (40 °C), screening a range of different templates that was available.
AT-Bph PCR (left), DR-Bph PCR (right), Red circle - Desired band size
However, we did not have enough time to continue with the rest of the construction.
In conclusion, we learnt that:
- For PCR, the annealing temperature used should be determined by the annealing region of the primer. In this case, at the start, we have been using annealing temperatures that were too high for the annealing region. As such, only when a lower annealing temperature was used (40 °C) did a good amplification occur.
- A low annealing temperature, however, increases the chances of non-specific amplification, which can be seen in the gel photos. To deal with this, new primers should be designed, where the annealing region to the template is increased, while reducing the upstream region of the restriction cut site.
- For plasmid backbones cut with compatible ends, alkaline phosphatase treated products used should be made fresh each time, to reduce the chances of re-ligation.
Heme Oxygenase 1
The PCR optimisation of the RBS-coupled heme oxygenase 1 coding sequence was successful from the start (see notebook), and much more effort was put into the construction of the HO-BioBrick, and the assembly with a T7 promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005 BBa_I719005] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I712074 BBa_I712074]).
After the initial failure of the ligation of the HO-PCR product with the plasmid backbone, and the lack of specificity of the colony PCR (see notebook), the BioBrick construction pipeline was adopted, using a new batch of alkaline phosphatase treated backbone, and the XbaI restriction digest as a screening method. Despite getting only 1 colony from the transformation, the single colony proved to be right on target, giving us our first successful BioBrick.
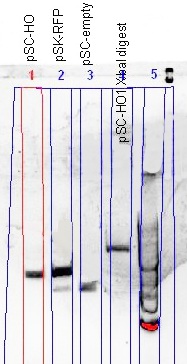
(Left) The comparison of the uncut plasmid size (pSC-HO) with the RFP-plasmid (pSC-RFP) and the re-ligated plasmid (pSC-empty) showed a successful insertion of the HO gene; smaller than RFP-plasmid, but larger than the re-ligated plasmid. The XbaI digest gave prove of the gene orientation; a plasmid able to be digested with XbaI.
(Right) Confirmation of the HO-BioBrick ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K646000 BBa_K646000]) was performed, using both PCR and a restriction digest using XbaI and PstI. Although the gel was not well resolved, the similarity of the PCR product size when compared with the original template, and that a gene fragment was cleaved out of the HO-BioBrick, was good enough to conclude that the HO-BioBrick construction was successful.
To test our HO-BioBrick, we assembled it to a T7 promoter, supplied in the iGEM distribution pack (plate 1, location 15N and 6N - Parts BBa_I719005 and BBa_I712074). Using our alternative assembly method, we digested the T7 promoter bricks with SpeI and PstI, and inserted our HO-BioBrick cut with XbaI and PstI, transformed into Bl21(DE3)Tuner E. coli cells. Selection was done on ampicillin plates (T7 bricks have amp-resistance) that also contained ALA (d-aminolevulinic acid, for heme pathway) and IPTG for induction of protein expression in Bl21(DE3)Tuner E. coli cells. A functional heme oxygenase would be able to metabolise heme into biliverdin, which causes the cells to look green (colour of biliverdin).
Liquid media containing IPTG and ALA, incubated for 24 hours at low temperature, spun down. Pellets are green as heme oxyganse was expressed using T7 promoter and degraded heme into biliverdin.
Conclusion
We probably need a conclusion.
 "
"