Team:UT-Tokyo/Project/System
From 2011.igem.org
System

iGEM UT-Tokyo
Project Overview
Our project aims at improving the working efficiency of bioremediation by increasing bacterial density near the substrate. To do this, we developed two types of E. coli, “guiders” and “workers”. The guiders are able to detect the substrate, and attract workers to themselves, where there is a high substrate concentration, when they do so. When the workers arrive around the substrate, they perform the bioremediation task assigned to them. The workers gather by chemoattraction and are then prevented from diffusing by a cell arrest mechanism, making the high density stable and last.
Chemotattraction
E.coli natively have a property called 'chemotaxis,' and attracted to certain substances, 'chemoattractant.’ E.coli sense concentration gradient of the substances and advance in the direction where the concentration is higher.
In this part we use chemoattraction to raise cell density. Specifically, we aim to realize substrate responsive chemoattrtactant overproduction and following cell assembly around the substrate.
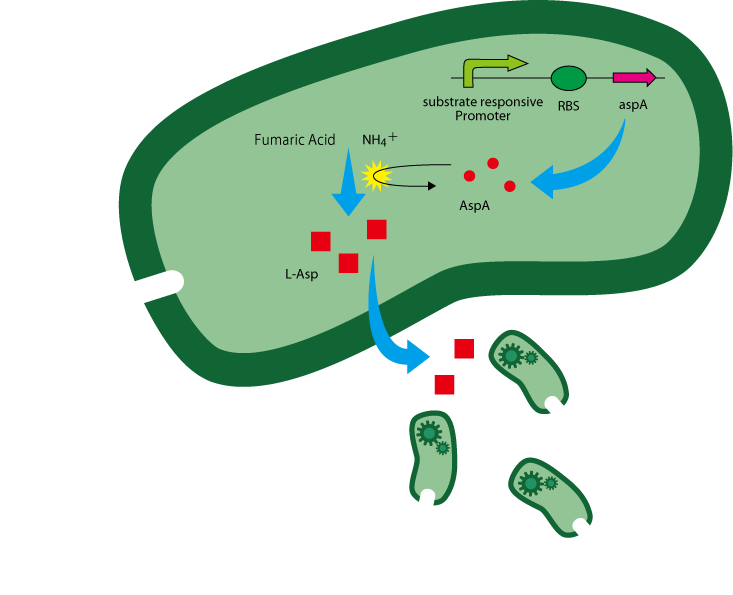
We have utilized L-Aspartate(L-Asp) as a chemoattractant. L-Asp is an amino acid verified as chemoattractant through preceding study[ref.1] and our own experiment. By making E.coli discharge L-Asp, L-Asp gradient around the cell is generated and other E.coli (workers and other guiders) are attracted and gather toward the cell, and higher cell density is created around E.coli secreting L-Asp.
Next, we have devised substrate responsive L-Asp overproduction mechanism. If guiders overproduce and secrete L-Asp in response to a particular substrate, they attract other bacteria and raise cell density around the substrate.
To make E.coli overproduce L-Asp when they sense a substrate, we attempt to insert the coding sequence of aspA in the downstream of a promoter responsive to the substrate.
Pres - rbs - aspA - d.Ter
The reaction of L-Asp production from fumaric acid and anmonium ion is catalyzed by aspartase(AspA) encoded by aspA. [ref.2][fig.1].
The gene aspA is derived from E.coli and L-Asp is overproduced in the presence of enough AspA and the two substrates above.
Once guiders sense a substrate and generate L-Asp gradient, other E.coli gather around the substrate as the result of chemoattraction. Among them, guiders attracted to the substrate spot also overproduce L-Asp if they sense the substrate. Here a positive feedback loop to create and keep gradient of L-Asp is formed. [fig.2]
Cell Arrest
Aspartate concentration gradient produced by guiders is gradually lost by diffusion as the time elapses. Therefore, in order to maintain the high worker bacteria density achieved by aspartate-dependent chemotaxis, it is required for the worker to be 'arrested' in the target location. To achive this, we have developed a mechanism that make bacteria less motile when cells are in target location by manipulating flagellum movement when they sense the substrate.
Bacterial chemotaxis and flagellar movement has been extensively investigated and the molecular basis for E.coli chemotaxis has already been elucidated. According to previous study[1] and our experimental result, a gene named cheZ, which codes for protein involved in chemotactic signaling pathway, regulates cell motility. More specifically, cheZ gene in E.coli genomic DNA is constitutively expressed in physiological condition, causing the cell to become motile by affecting a flagellar motor binding protein called CheY. Therefore, cells are motile in the presence of cheZ and immotile in the absence of cheZ. (Learn more about chemotaxis pathway in E.coli)
In order to make "worker" bacteria stay at the target site, we need to lower the cell motility in response to the substrate. Put in another way, we want the expression of cheZ to be activated in the absense of substrate and repressed in the presence of substrate.
To achieve this, we utilize cheZ knockout strain of E.coli to eliminate baseline expression and devised the gene construct shown below, in which cheZ expression is repressed in a substrate-dependent manner (fig. 2).
cIp -rbs - CheZ - d.Ter - Pi - rbs - cI - d.Ter
When the cell is outside of the target location, cI inhibitor is not expressed, whereas CheZ is expressed and the expression product rescues motility of CheZ- strain. Once the cell gets inside the target location, substrate-responsive promoter is activated and cI inhibitor is expressed. cI inhibitor then inhibits the activity of cI promotor and thereby represses CheZ expression, which leads to substrate-induced loss of motility.
 "
"