Team:Harvard/Technology/Chip Synthesis
From 2011.igem.org
MAGE | Lambda Red| Chip-Based Library | Protocols
Chip-Based DNA Synthesis
We are creating 55,000 zinc fingers using microchip synthesis (Kosuri et al). These fingers will then be tried against the DNA sequences we wish to bind.
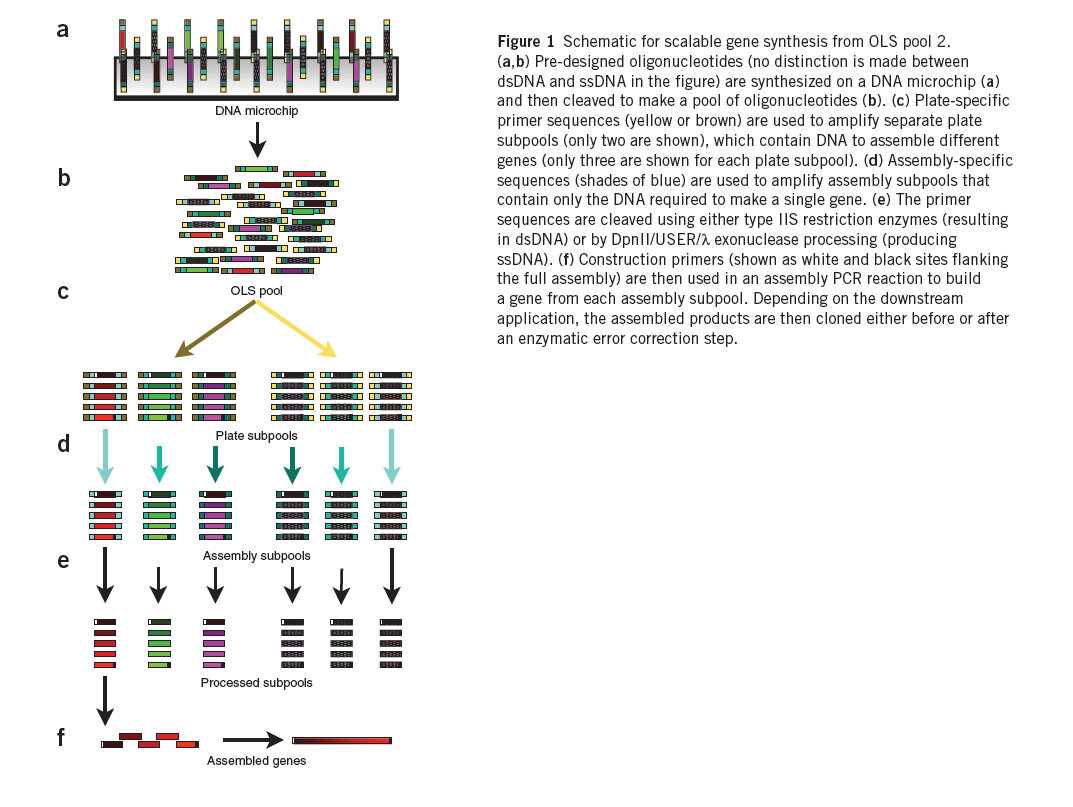
Summary (Adapted from Kosuri et al)1
The synthesis of DNA encoding regulatory elements, genes, pathways and entire genomes provides powerful ways to both test biological hypotheses and harness biology for our use. For example, from the use of oligonucleotides in deciphering the genetic code to the recent complete synthesis of a viable bacterial genome, DNA synthesis has engendered tremendous progress in biology. Currently, almost all DNA synthesis relies on the use of phosphoramidite chemistry on controlled-pore glass (CPG) substrates. The synthesis of gene-sized fragments (500–5,000 base pairs (bp)) relies on assembling many CPG oligonucleotides together using a variety of gene synthesis techniques. Technologies to assemble verified gene-sized fragments into much larger synthetic constructs are now fairly mature.
The price of gene synthesis has fallen drastically over the last decade. However, the current commercial price of gene synthesis, ~$0.40–1.00/bp, has begun to approach the relatively stable cost of the CPG oligonucleotide precursors (~$0.10–0.20/bp)1, suggesting that oligonucleotide cost is limiting. At these prices, the construction of large gene libraries and synthetic genomes is out of reach to most.
A promising route is to harness existing DNA microchips, which can produce up to a million different oligonucleotides on a single chip, as a source of DNA. Recently, the quality of microchip-synthesized oligonucleotides was improved by controlling depurination during the synthesis process. These arrays produce up to 55,000 200-mer oligonucleotides on a single chip and are sold as a ~1–10 picomole pools of oligonucleotides, termed OLS pools (oligo library synthesis). Estimations of the frequency of transitions, transversions, insertions and deletions in OLS pools found the overall error rate to be ~1/500 bp both before and after PCR amplification, suggesting that OLS pools can be used for accurate large-scale gene synthesis.
Protocol
qPCR of subpools
Complete protocol found in the supplementary material of Kosuri 2010
Perform one qPCR reaction for each single stranded sub-pool, using the following recipe:
- 100 ul Sybr Green master mix
- 200 nm primer
- 2 ul pooled oligo
- Water to bring total volume up to 200 ul
When performing the PCR reaction, one wants to watch the graphs produced by the thermocycler. Initially, the reaction will proceed at an exponential rate. However, as soon as it begins to plateau, the reaction should be stopped. Until this point, oligos were being amplified equally. However, once the plateau is reached, oligos are amplified at unqual rates. It will take approximately 10 cycles to reach the plateau.
On the thermocycler, run the following program:
- 1 minute at 95*
- 10 seconds at 95*
- 30 seconds at the primer annealing temperature
- Go to step 2 until the reaction plateaus.
The PCR product should undergo PCR purification.
Digestion of oligos with Type II Restriction Enzyme
Digesting the oligos with a type II restriction enzyme will result in sticky ends that can anneal to the backbone during the ligation step.
Following the reaction, the oligos should undergo a PCR cleanup.
Double digestion of backbone
A double digestion on the backbone decreases the likelihood of the backbone self-ligating. When designing the backbone, a Type I cut site was inserted in the cut out gap that resulted from digestion with the Type II restriction enzyme. If the cut out gap is still present on the plasmid (as is the case if the digestion is not completely successful), then the Type I restriction enzyme with cut the plasmid, resulting in blunt ends that are less likely to self-ligate.
Following digestion, the backbone undergoes a phosphatase reaction for an hour, to decrease the likelihood of self-ligation by removing the 5’ phosphate.
Following the phosphatase reaction, the backbone should undergo a PCR cleanup.
Ligation of oligo library and backbone
When ligating 1000 ng of backbone, and insert:backbone ratio of 2:1 is optimal for our reactions. Prior to adding the ligase, the reaction is heated to 37* for 5 minutes, to prevent any non-specific annealing. Once the reaction cools back down to room temperature, the ligase is added, and the reaction is allowed to run for 10 minutes at room temperature.
The ligation product should be run through a min-elute column, taking care to elute in 10 ul of water.
Transformation
100 ng of ligation product should be transformed into library competent cells.
</div>
 "
"