Team:DTU-Denmark/Regulatory mechanisms
From 2011.igem.org
What is: How do (trans) sRNAs work as regulators
How do (trans) sRNAs work as regulators
The key-word for sRNA mediated regulation is base-pairing. A part of the sRNA shows limited complementary to the mRNA of the gene it regulates and the sRNA can thus base-pair with the mRNA. The region of base-pairing is typically only 6-20 nucleotides, and often it is only a subset of these nucleotides which seems to be critical for regulation[2][3]. This (often imperfect) base-pairing between the sRNA and target mRNA leads to changes in mRNA translation or stability - or both, and thereby influences the target gene expression[1]. sRNAs can be both activators and repressors for gene expression depending on what part of the mRNA molecule they base-pair with.
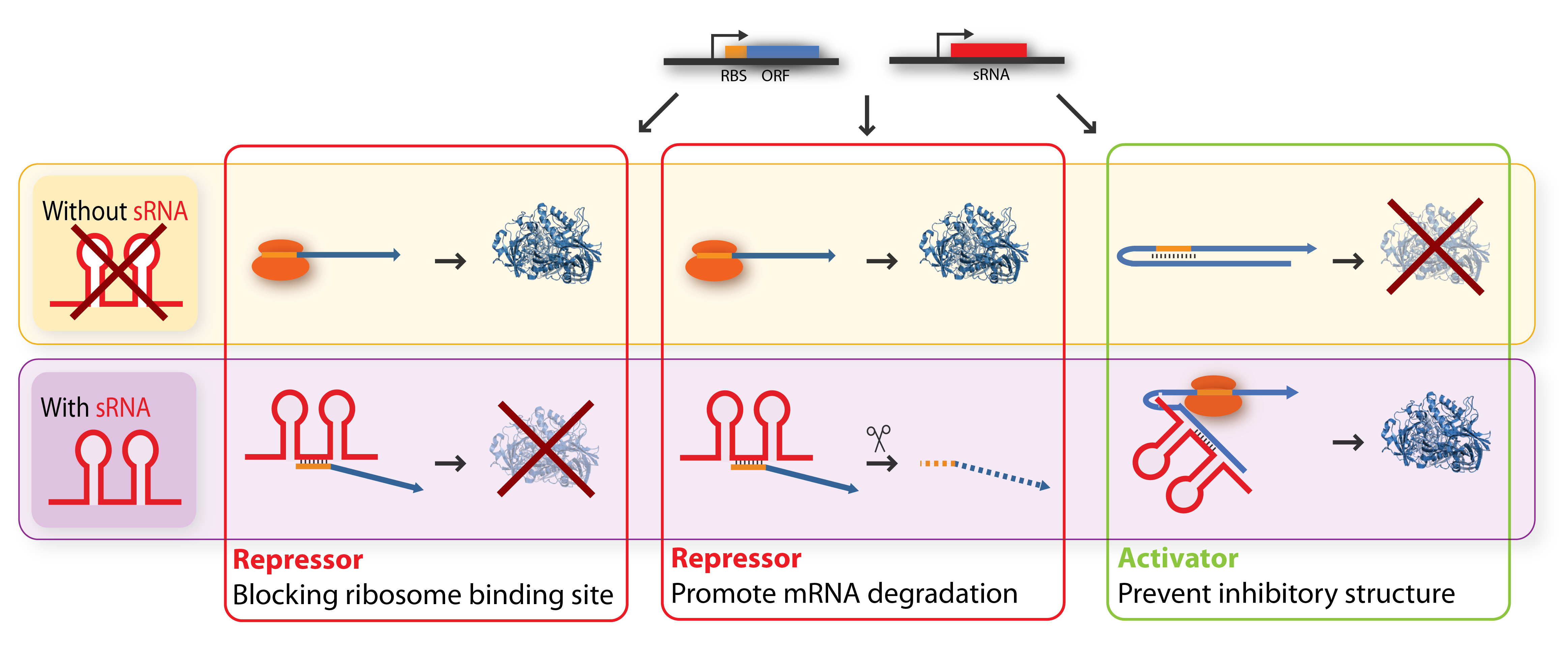
Repressor sRNAs act negatively by binding to the 5’ UTR often near the ribosome binding site. The binding inhibits translation by impeding ribosome binding and/or target the mRNA for degradation by RNases (often RNase E). Note that the degradation entails irreversible regulation[3]. Some sRNAs act stoichiometrically meaning that they are co-degradated with the mRNA whereas others act catalytically and are not degraded in the reaction.
Activator sRNAs act positively through an anti-antisense mechanism where sRNA base-pairing with the target mRNA disrupt an inhibitory secondary structure sequestering the ribosome-binding site. As a result, the ribosome-binding site is liberated and free to bind ribosomes[3].
References
[1] Beisel, Chase L., and Gisela Storz. “Base pairing small RNAs and their roles in global regulatory networks.” FEMS Microbiology Reviews 34, no. 5 (2010): 866-882.
[2] van Vliet, Arnoud Hm, and Brendan W Wren. “New levels of sophistication in the transcriptional landscape of bacteria.” Genome biology. 10, no. 8 (2009).
[3] Waters, Lauren S., and Gisela Storz. “Regulatory RNAs in Bacteria.” Cell 136, no. 4 (2009): 615-628.
 "
"








