Team:Paris Liliane Bettencourt/Notebook/2011/09/15/
From 2011.igem.org

Contents |
Baptiste, Hovannes & Ouriel
Microscopy experiments of today.
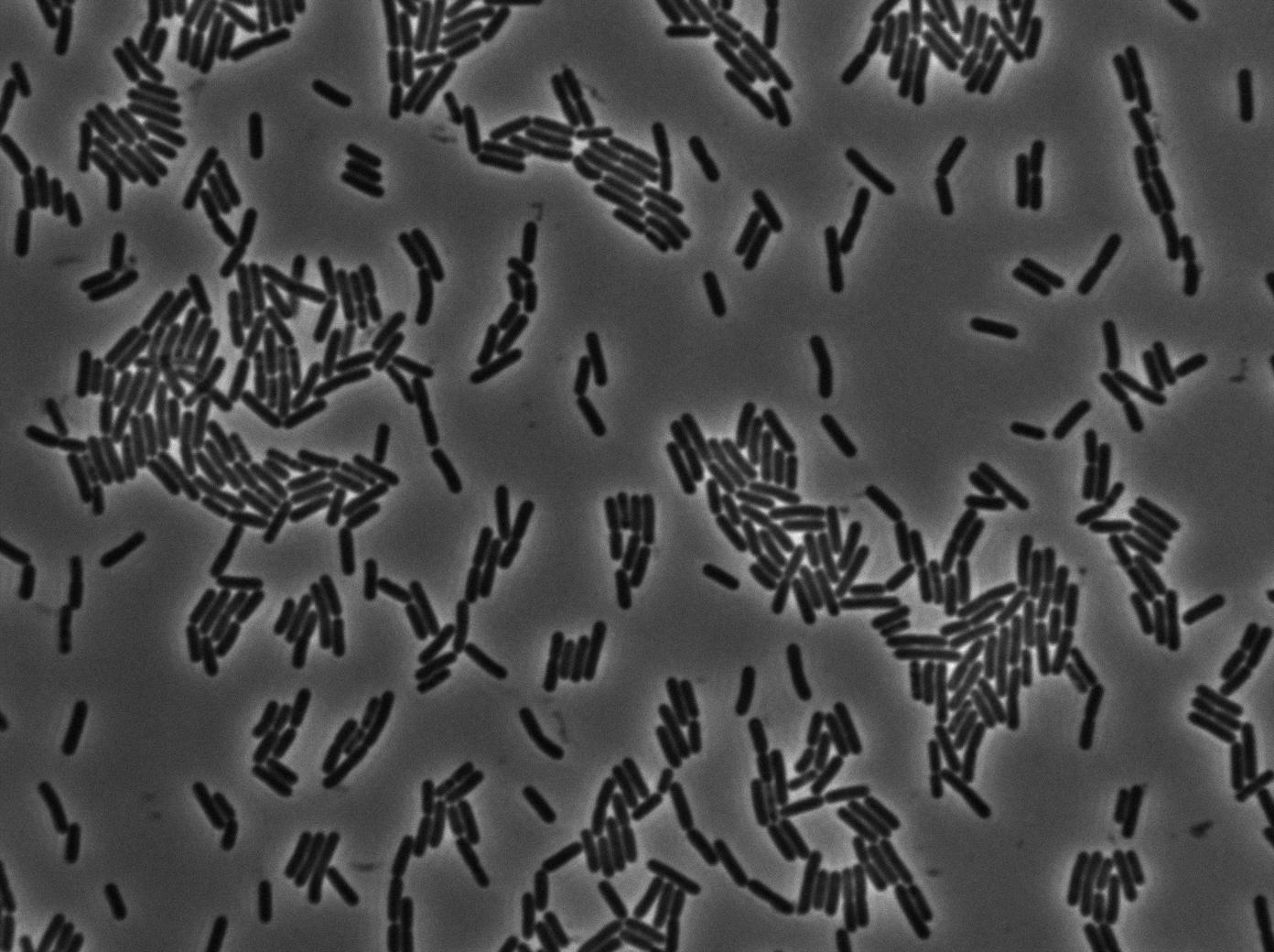
As we begin to master totally the Nikon microscope we chose to run three settings for the 3610 gfp-/3610 gfp+ experiments. The aim is to see which is the best way to plate the cells on our slides and more importantly at which growth state. We started from the same overnight culture and then created three microscopic slides from our two 3610 strains (gfp+ and gfp-):
- One directly from the overnight culture in stationnary phase.
- One with dilution until the OD reached 0.1. We then waited for the OD to grow to 0.8 (roughly 2 hours) and then concentrated the cells (4000 rpm during 10 min and then put 200 microliters of LB on the pellet).
- One by diluting the overnight culture in the ratio 1/500. We then waited for the OD to grow to 0.8 (roughly 4 hours) and then concentrated the cells (4000 rpm during 10 min and then put 200 microliters of LB on the pellet).
First conclusions of our methods
The autofocus programme on the Nikon was not perfect so it was adjusted to our needs. It gave us slightly blurry image for the second serie of slide (dilution to an OD of 0.1) and bad results for the stationnary phase slide. Fortunately, the stationnary phase means that bacteria do not grow so we were able to interpret the results even after missing more than an hour of images. The slide for the 1/500 dilution was very faulty. We could not see anything clearly and we will have to retry it tomorrow. To let the drop of cells dry or not, is the primary issue here. Both ways will be tried tomorrow to see which one is suitable.
Stationnary phase
As expected, no growth and no diffusion was observed.
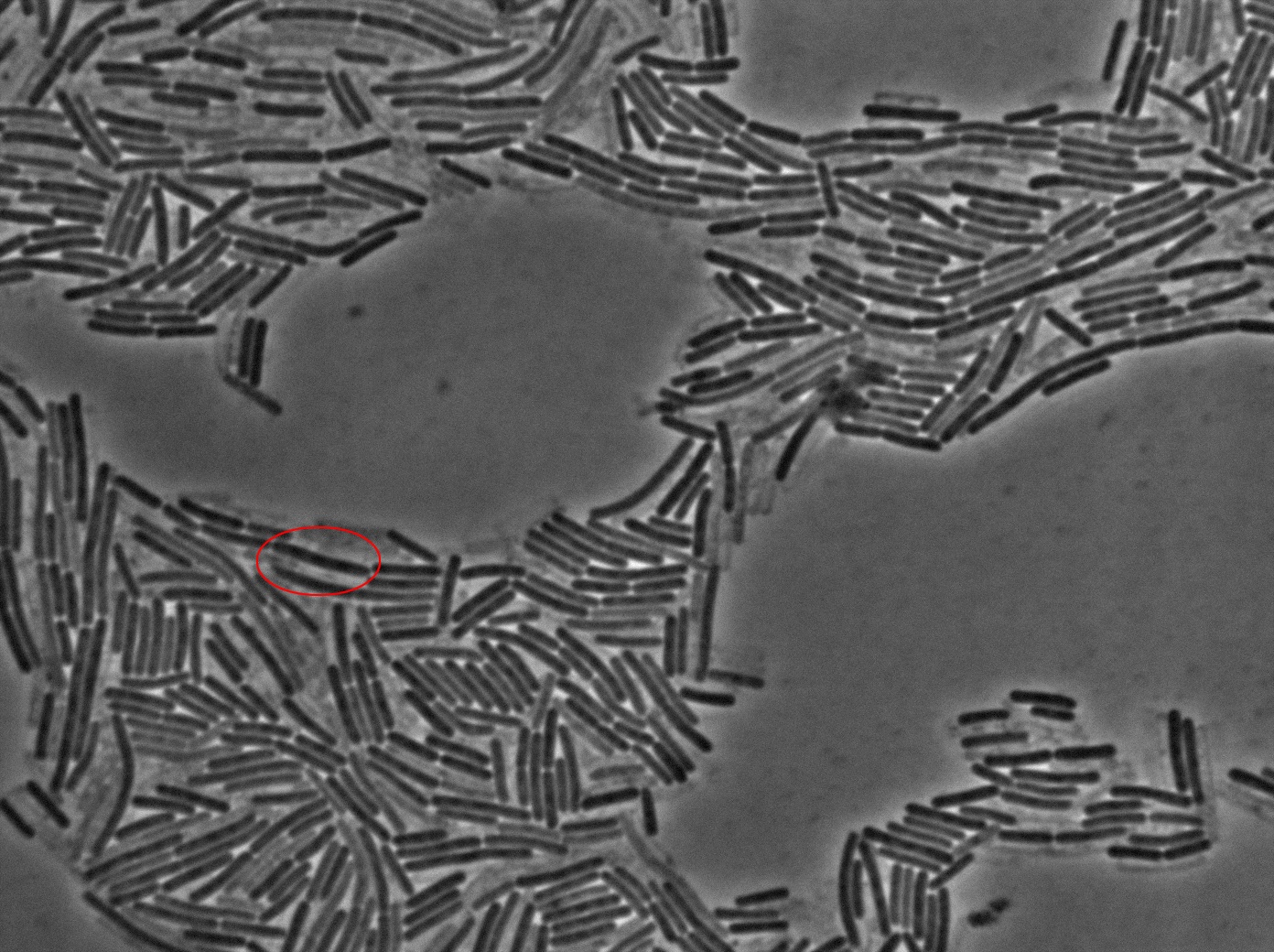
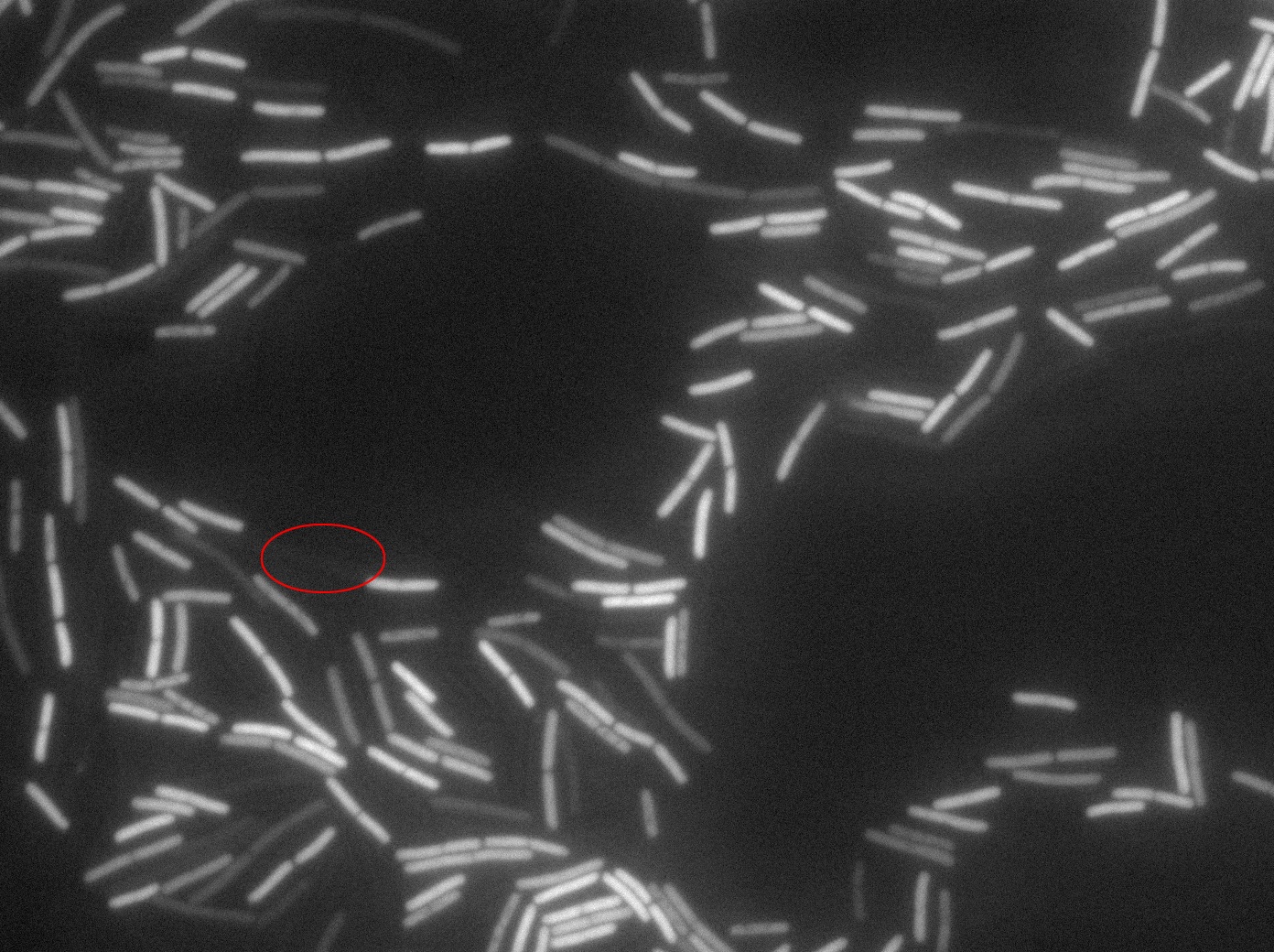
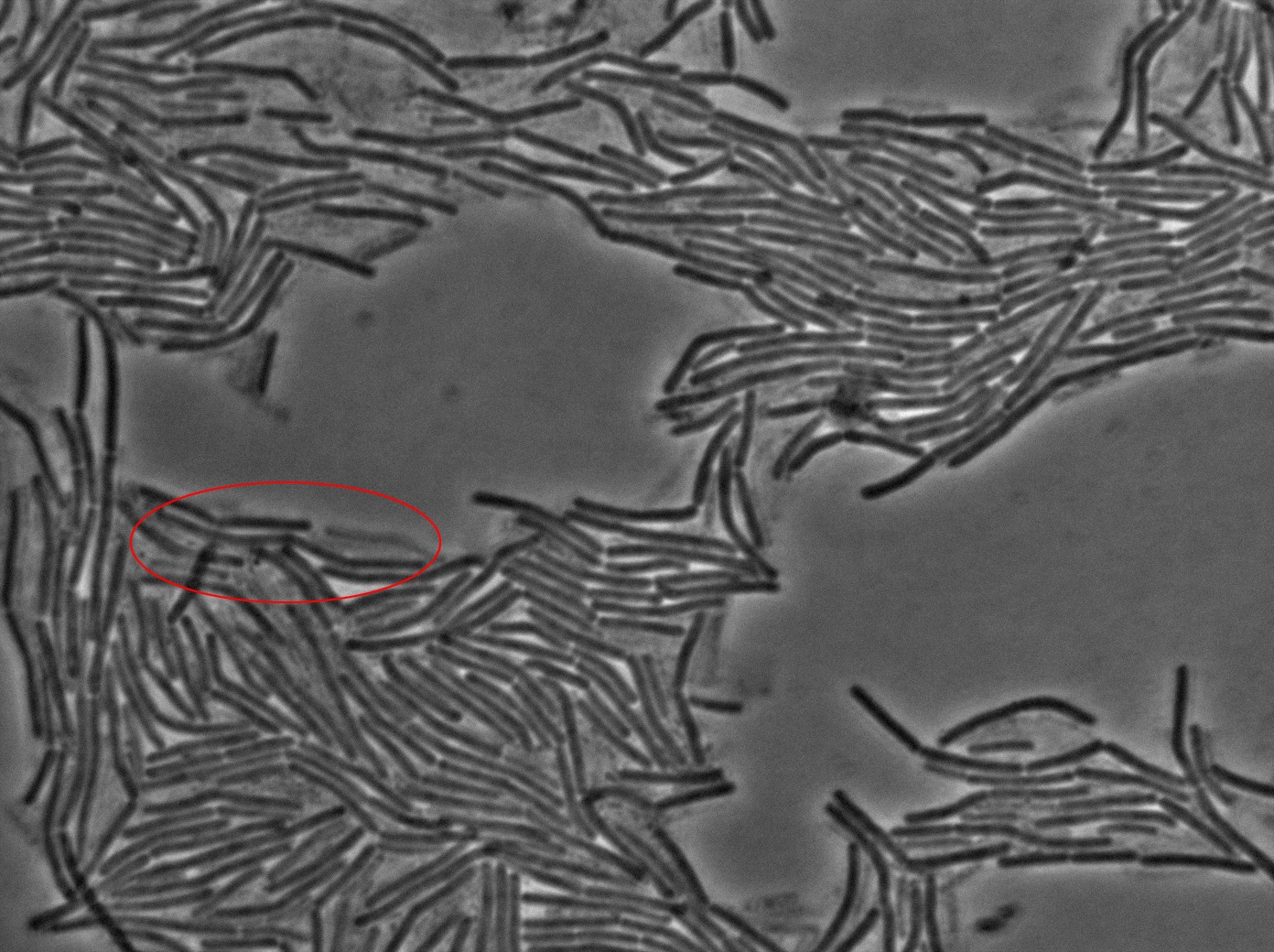
Dilution to an OD of 0.1
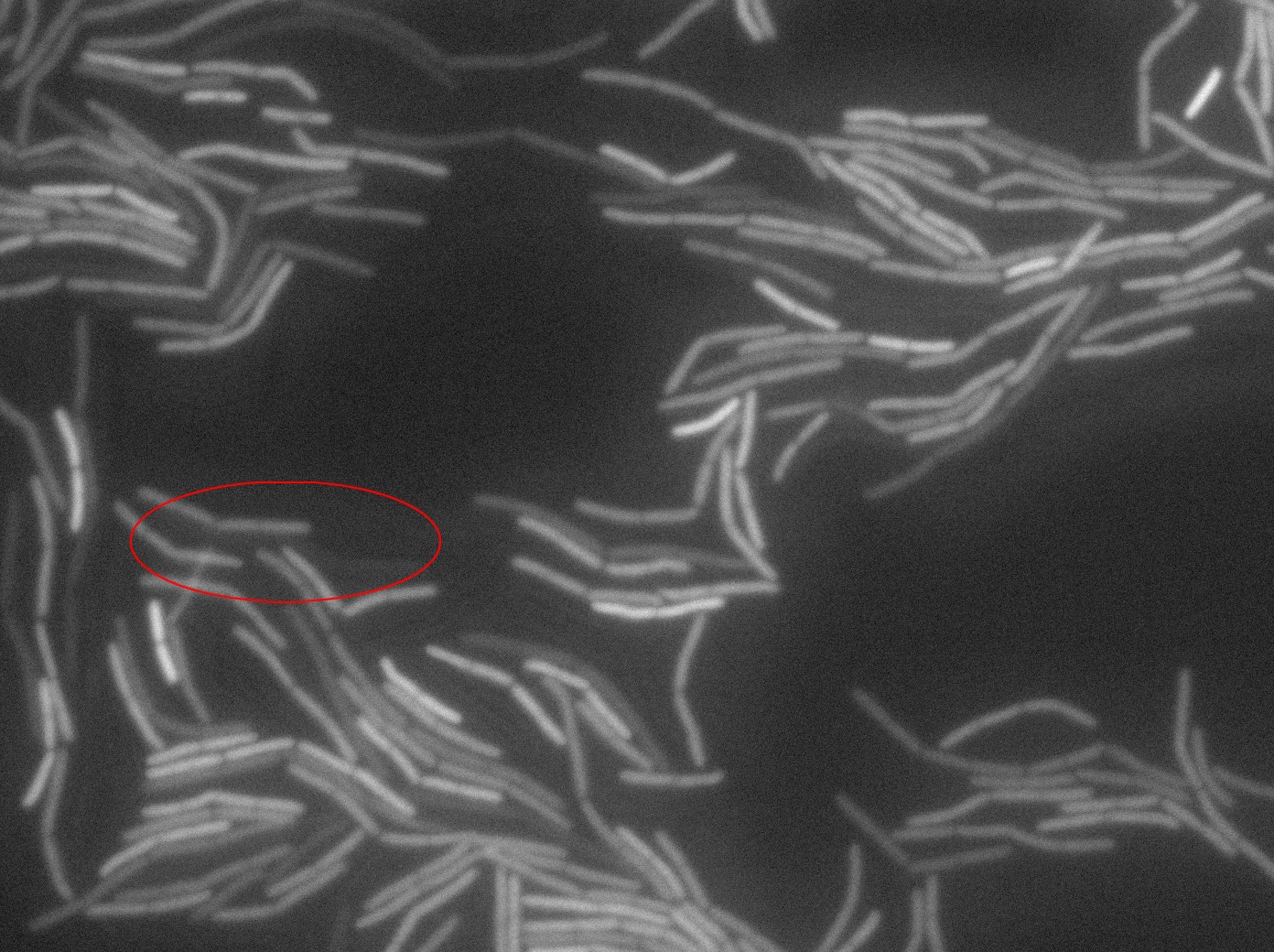
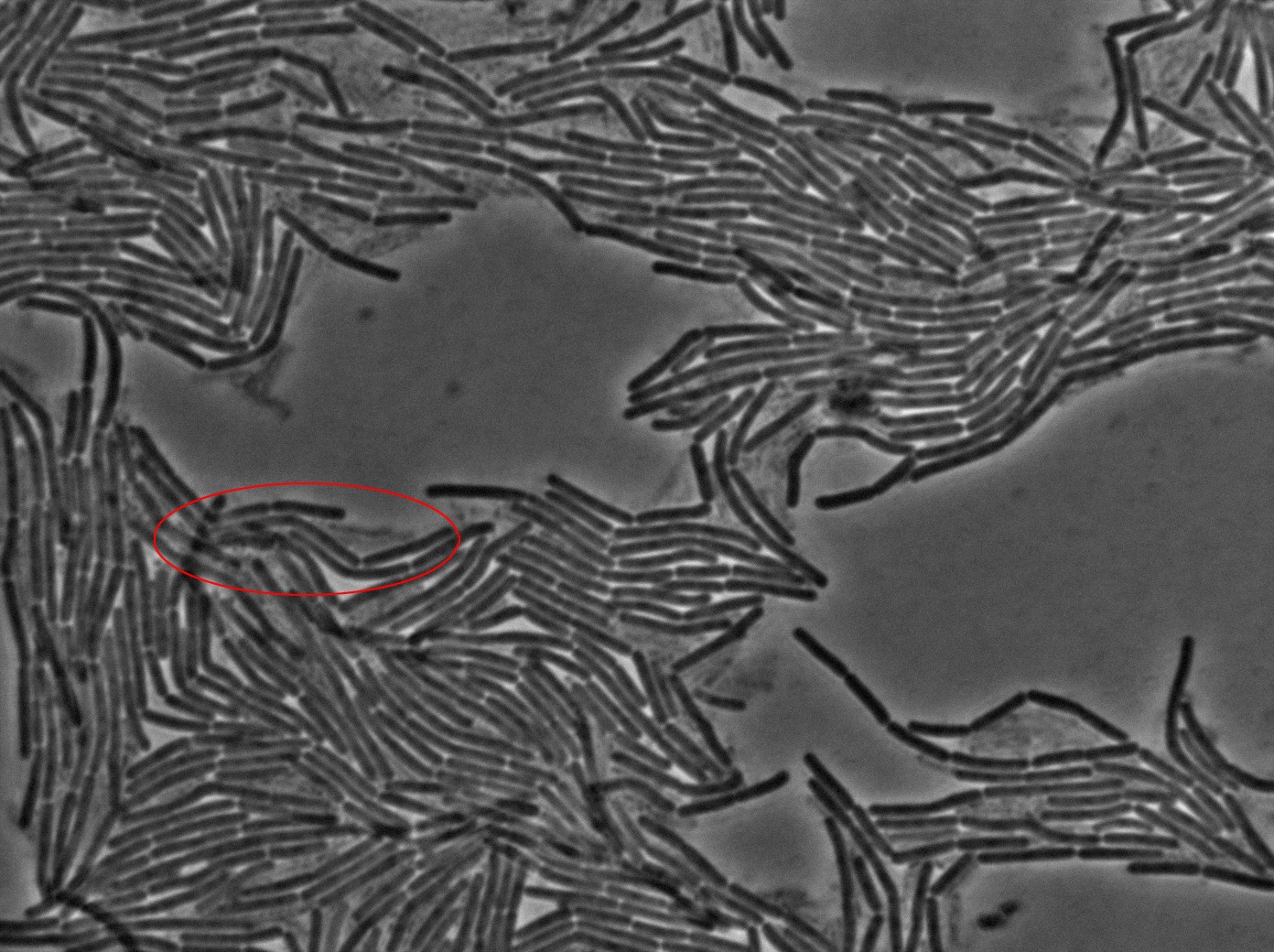
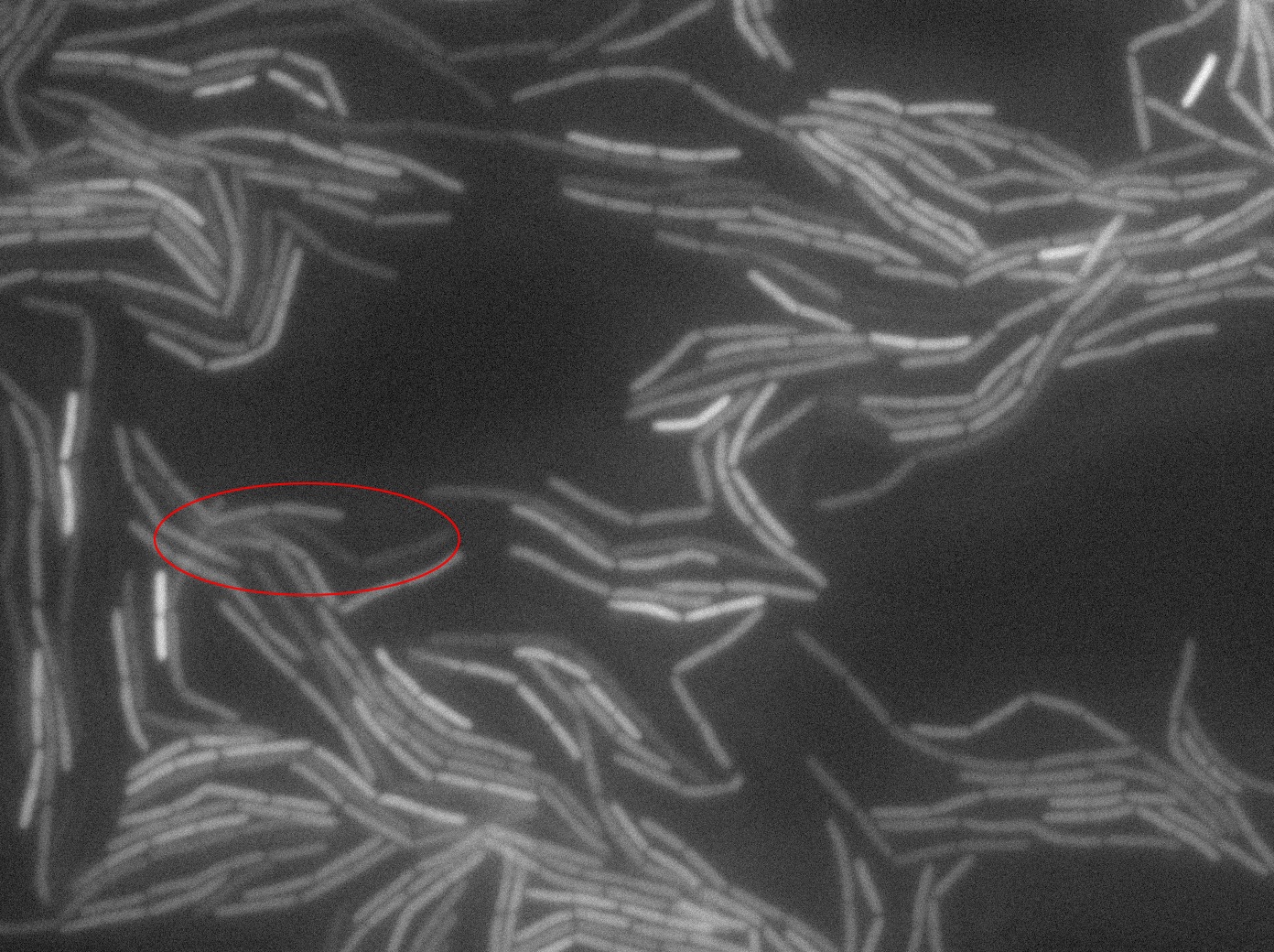
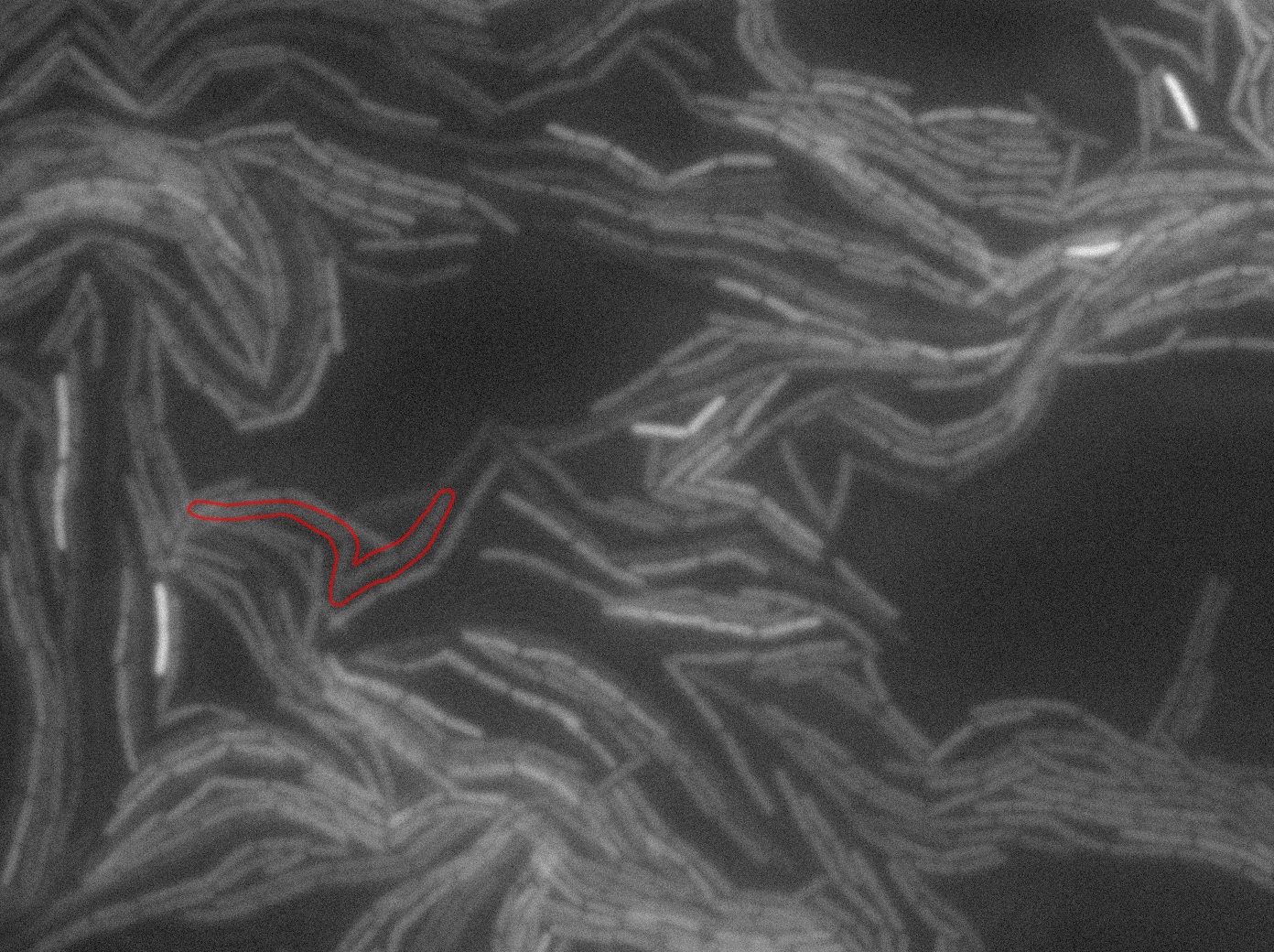
The slide was one of the best we've done so far. We were able to see clearly zones with a monolayer of roughly 50/50 GFP/non-GFP cells. We finally found some evidence of GFP diffusion in one of the followed regions of the slide! The cells gaining fluorescence are circled in red. Note that, gfp+ cells lost their fluorescence with time whilst gfp- (the ones circled in red) gained fluorescence. We also run positive controls but took pictures only at the end of the experiment.
Kevin
Following step for characterisation of tetO array
Double transformation works ! Tomorrow microscopy.
cyrille
Treatment of the ligations done yesterday. PVeg T7a TT + pT7 gfp TT worked. 16 col putted in culture.
For the other ligations, the results of the positive control indicates that the transformation failed. 1 clone was found for p/eg tRNA rfp, and 3 pVeg tRNA tt rfp they were insulated. PCR colonie failed. The cultures will be minkprepped tomorow.
The ligation that are supposed to have failed were done again. As we were short on pHM3, the S99 was not done again.
Miniprep of pHM3 overday.
Suspicion on pHM3 on the lost of replication origin for subtilis due to the analysis of the sequencing. 7 miniprep of the original pHM3 were launched. Double digestion in HincIII and PstI was help to see if the region was indeed lost. HindIII cut several times in the replication origin region.
 "
"