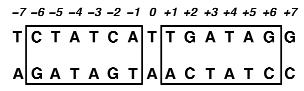Team:EPF-Lausanne/Our Project/TetR mutants
From 2011.igem.org
TetR mutants
to write here: how we got TetR mutants, with which method (we can also explain the method in the "tools" section) The MITOMI characterization will be explained in the "tool" section?
Background information about TetR
Widespread among bacteria, TetR is a repressor that regulates enzymes essential for resistance against the antibiotic tetracycline. These enzymes are encoded on the tet operon (TetO), which is repressed by TetR in normal conditions. When tetracycline is present, the antibiotic molecule binds to TetR and inactivates it, thus allowing the expression of TetO.
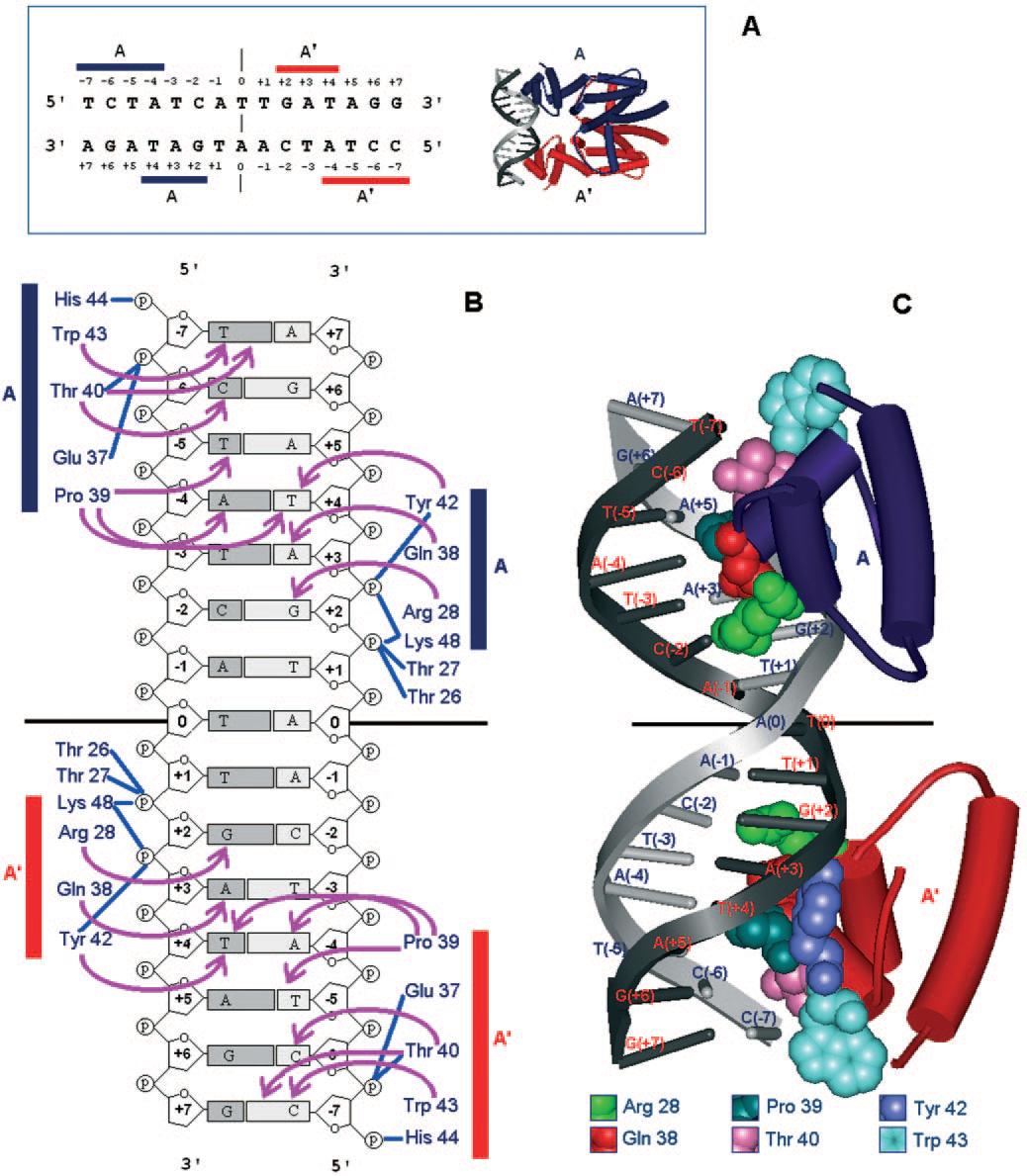
TetR forms a dimer, each part of the dimer being involved in DNA binding. Consequently, the recognition sequence of TetR is symmetrical (the two boxes on the image below), with a base pair separing the two sub-sequences. [http://www.nature.com/doifinder/10.1038/73324 Orth et al, 2000] The whole promoter, comprising this recognition sequence, is called Ptet.
The binding of each monomer to the recognition sequence has been studied toroughly; we know which amino acid interacts to which nucleotide, as you can see in the figure below. The amino acids directly involved in binding to the DNA are between position 26 and 48 of each monomer. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1197418/?tool=pubmed Ramos et al, 2005]
The impact of one or several amino acids changes on the binding strength and specificity is less known. Amino acids in direct proximity to or the residues binding DNA are probably crucial - but more distant amino acids could also have an influence due to allosteric effects. Some TetR mutants that have an altered recognition sequence have been characterized in litterature.
Lilia, please add a list... I have 4C, 6C (Helbl) and double from Krueger
 "
"