Team:NTNU Trondheim/Data
From 2011.igem.org

Data
rrnB P1 promoter
The rrnB P1 promoter is negatively regulated by ppGpp [Kilde] and therefore sutible for our stress sensor. We found the promoter in the registry distribution (BBa_K112118) submitted by Berkly in 2008.
This part is in the BBb standard form [http://openwetware.org/wiki/Template:AndersonLab:BBb_Standard] and is not compatible with the parts in the BBa standard. In order to get the part in BBa standard we PCR amplified the promoter using the BBa_K112118 as template and primers containing the BBa prefix and suffix:
| Primer | Type | Sequence |
|---|---|---|
| rrnB P1 F | Forward | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGACGTATCCTACGCCCGTGGT |
| rrnB P1 R | Reverse | GTTTCTTCCTGCAGCGGCCGCTACTAGTACGCCTTCCCGCTACAGAGTCA |
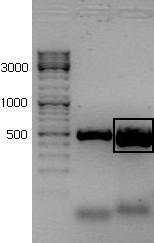
The PCR-product was run on a 1,5% agarose gel in order to verify that we had achieved the correct product and to separate it from any other unwanted products.
The new rrnB P1 biobrick was inserted in the psb1C3 plasmid provided by the iGem HQ. Plasmid from five colonies were digested with the enzymes BstBI and SpeI It seems like parallel 1 and 2 have the insert.
Stress Sensor
The stress sensor consist of MCherry (a RFP gene) controlled by the Lac promoter and the Lac inhibitor (LacI) controlled by the rrnB P1 promoter. The MCherry biobrick (BBa_J06702) is complete with both RBS and a terminator sequence, and we fist connected this part with the LacP biobrick BBa_R0011. The LacI biobrick BBa_C0012 had a RBS region but no terminator, so we added the term biobrick BBa_B0015. Then we connected these to constructs to get the final stress sensor minus the P1 promoter. In order the check if the construct was correct we digested the plasmid with ........
Then we added the P1 promoter using the PCR-product as insert. To check if the part was inserted we digested the resulting plasmids with BstBI cutting in the promoter region.
 "
"