Team:Missouri Miners/Project
From 2011.igem.org
Project
Abstract
In the bodies of people with diabetes, the ability to recognize and respond to glucose concentrations in the blood has been compromised. As a result, glucose accumulates to dangerous levels. High blood glucose concentrations can cause irreversible damage to critical organs, impairing their functionality. With parts from the iGEM registry, our team created a glucose-controlled promoter linked to a yellow fluorescence production gene in E. coli. The concentrations of glucose to which the promoter responds can be determined. Once the concentration is known, the promoter can be mutated so that it will be activated by varying concentrations of glucose and be used as a glucose sensor for people with diabetes. In the future, an insulin gene could be added to this system for use in insulin pumps, where specific glucose levels trigger insulin production in E. coli.
Our System Overview
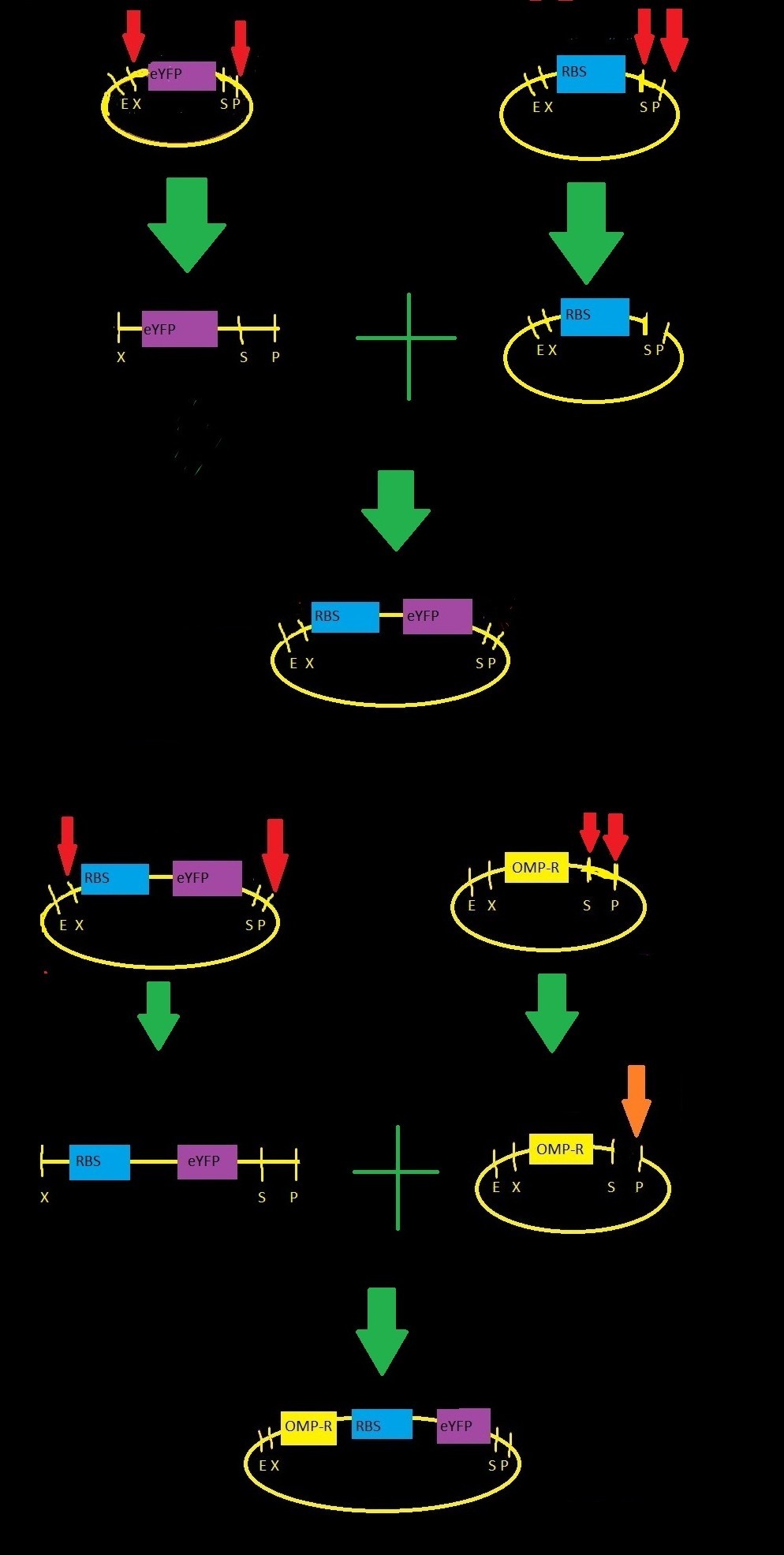
We submitted two biobricks to the registry: an intermediate part (http://partsregistry.org/wiki/index.php?title=Part:BBa_K621000) with a ribosome binding site and eYFP, and the intermediate plus the ompC operator (http://partsregistry.org/wiki/index.php?title=Part:BBa_K621001). The ompC operator has three binding sites for phosphorylated OmpR. OmpR and EnvZ work together in a two-component regulatory system as shown in the diagram below.
EnvZ is an inner membrane protein that senses osmolarity. Phosphorlyation of OmpR by EnvZ positively correlates with the osmolarity of the system. Phosphorylated OmpR (OmpR-P) can bind to the three sites on the ompC operator. When one or two of the binding sites are occupied by OmpR-P, RNA polymerase is recruited to begin downstream transcription of the reporter gene, eYFP. However, when all three OmpR binding sites are occupied by OmpR-P, RNA polymerase cannot bind, the reporter gene can no longer be produced, and therefore the system is inhibited. In summary, as osmolarity increases from very low levels, the fluorescence produced by the system also increases until the system reaches a threshold osmolarity. Once the system reaches the threshold, the fluorescence will decrease with increasing osmolarity due to the inherent down-regulation of the system. The activity of the system can be quantified using the fluorescence of the cells because the two-component regulatory system of EnvZ and OmpR regulates transcription of the eYFP gene, dictating the level of fluorescence.
Making Our Parts
We started with three parts from the iGEM registry:
- BBa_B0032: RBS-3, a medium-strength ribosome binding site
- BBa_E0030: eYFP gene, codes for yellow fluorescence
- BBa_R0082: omp-c operon, contains three binding sites for phosphorylated OmpR
Testing
To measure fluorescence our team used a 96-well plate reader. Overnight cultures of bacteria were grown in 0.5X LB media and then aliquoted into 1.5ml tubes. Serial dilutions of the desired glucose concentrations were made. Glucose was diluted using 0.5X LB media so that the concentration of LB remained constant. The glucose was administered to the cells in a fashion that did not dilute the cell densities in the cultures. Samples of each treatment were added to a 96-well plate and were imaged using the plate reader. See data page for results.
Next Steps
For our system to be able to sense smaller changes in osmolarity the OmpC operon needs to be mutated to a state of more sensitivity. This processes of mutating our system is called mutagenesis. How we do mutagenesis is by transforming our system into a mutagenic strain of E. coli where these cells have no proof reading capability during DNA replication. So the rate of mutations is increased by 1000x. Once we put our system into the the mutagenic strain we allow the cells to replicate in LB broth for overnight and then with the new mutagenic culture we do a plasmid prep to purify the mutagenic plasmids. Theoretically every single one of these plasmids are different. We then transform DH5α with the mutagenic plasmids and allow them to grow on LB amp plates overnight. Once the plates have decent colonies we now have to streak for isolation onto LB amp plates. Isolation allows us try to work with just one strain of our original system. From the isolation plate we will make LB broth cultures to then start our glucose trials at different concentrations of glucose. Glucose trials consist of making serial dilutions with glucose and LB broth. To quantify our results we will use a fluorescence measuring machine.
 "
"