Team:Paris Bettencourt/Experiments/T7 diffusion
From 2011.igem.org

T7 diffusion experiments
Design overview
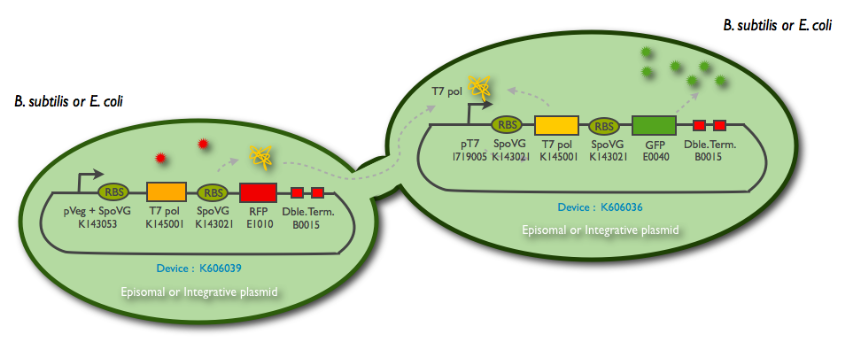
 Fig1: Schematic summary of the T7 diffusion device
Fig1: Schematic summary of the T7 diffusion device
More information on the design here : link
Parts
We synthetized the pHyperSpank promoter (K143055), but we were not able to incorporate it into our biggest construct. Instead, we cloned a Pveg promoter SpoVG(K143053) that has a constitutive expression.
The following schematic represents what we have indeed managed to clone:
This summary shows the parts we have characterized, and the ones we have sent to the registry. But for the moment, we didn't manage to clone them into an integration vector (K090403) nor a replicative vector (pHM3) for subtilis, as well as the combination of the emittor and the receiver inside the same plasmid.
Characterization of the T7 promoter
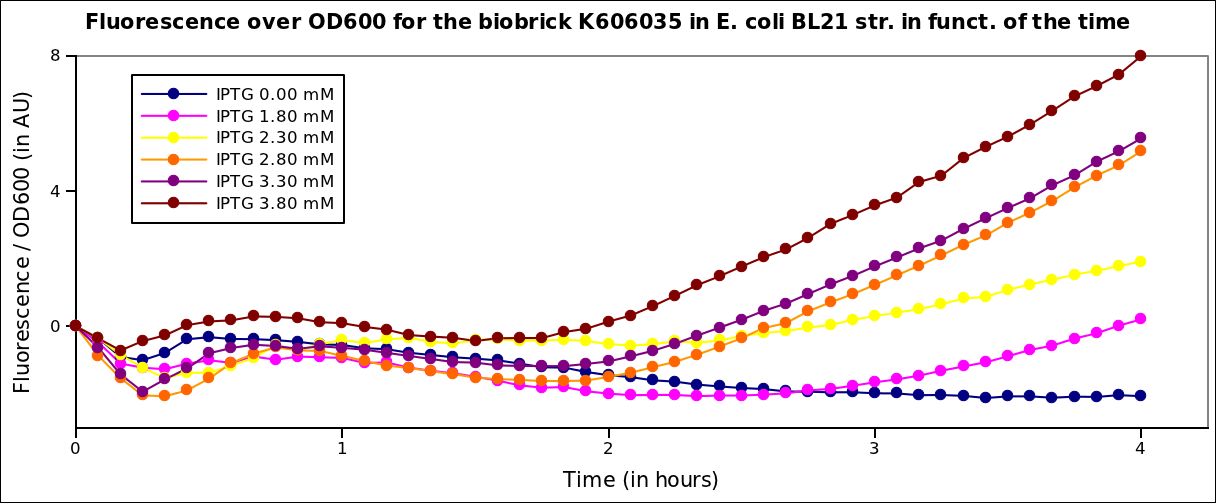
In order to characterize the pT7 promotor, we used the construct: pT7-RBS-GFP-T7ter. This construct was transformed into BL21 strains expressing the T7 polymerase under IPTG induction.Fluorescence kinetics
The measurements have been carried out on a spectrophotometer, at 37°C under transient shaking. The experiment lasted 4h, we tested several colonies and several IPTG concentrations. The OD 600nm and the fluorescence of the GFP (exc: 470nm / meas:515 nm) was measured every 5 min, and the ratio of the two was calculated.
All values were normalized by substracting the fluorescence/OD value of the well with 0 mM IPTG at time 0. The values given are in arbitrary units.
After 2 hrs of induction, we see a clear increase of the fluorescence proportional to the IPTG concentration (that is to say with the quantity of T7 polymerase induced in the cell). After 4 hrs, the expression of GFP under the pT7 is still not saturated
Here, we plot the ratio of induction of the T7 polymerase dependant construct for the different concentrations of IPTG at a given time (4 hrs), taking the well with 0 IPTG at time 0 as the reference.
Characterization of the T7 signal amplification leakage in E. coli
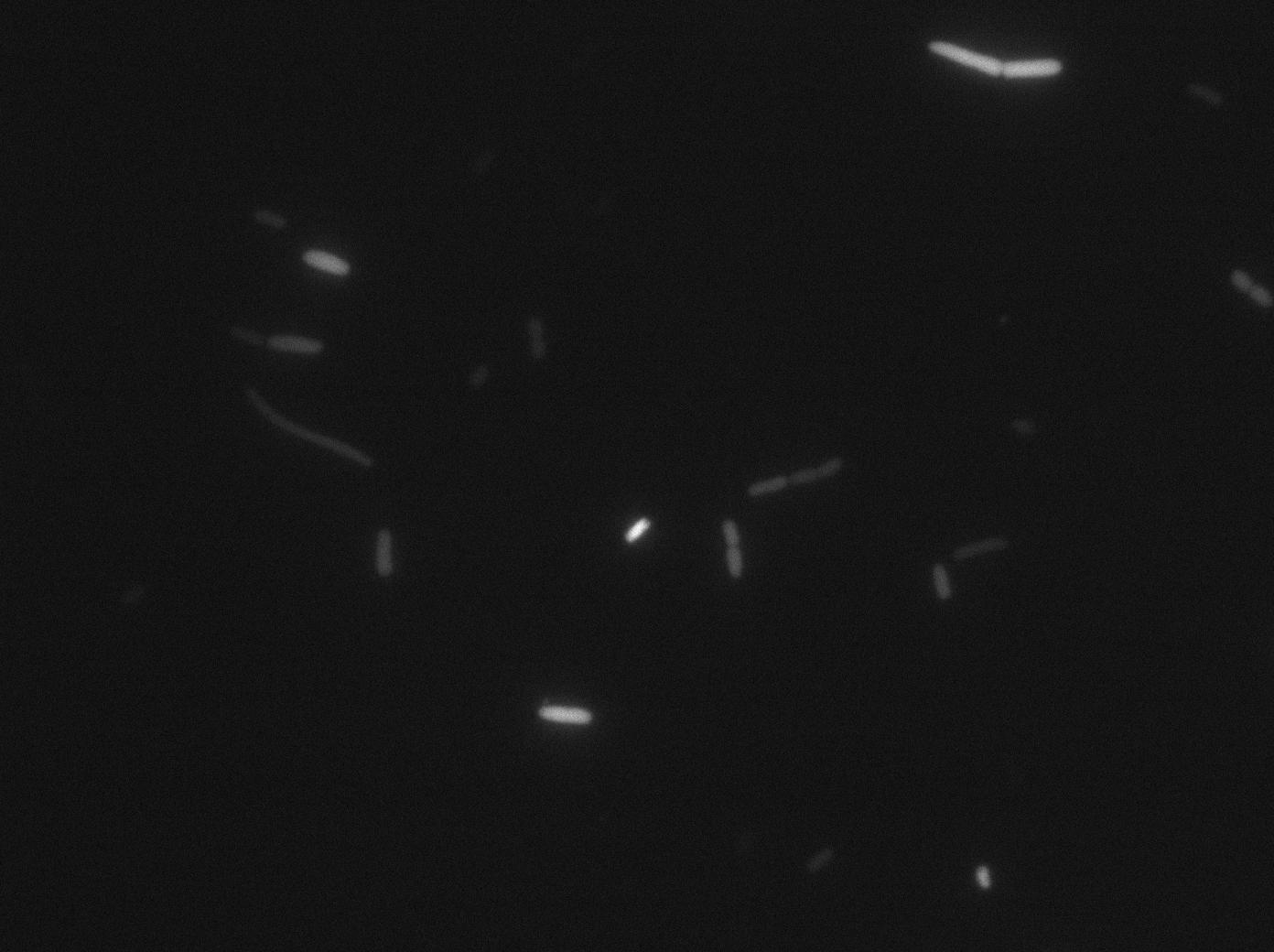
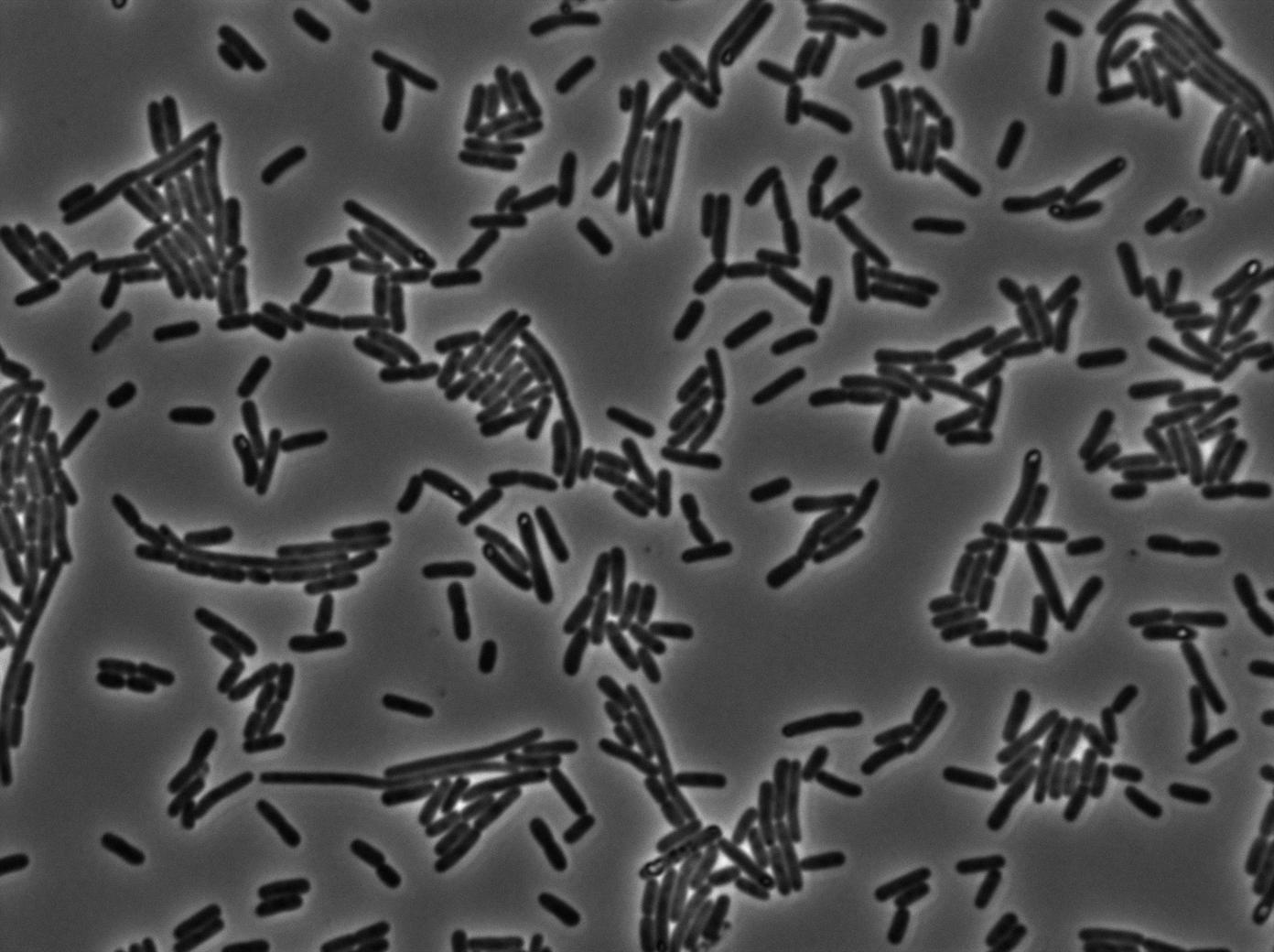
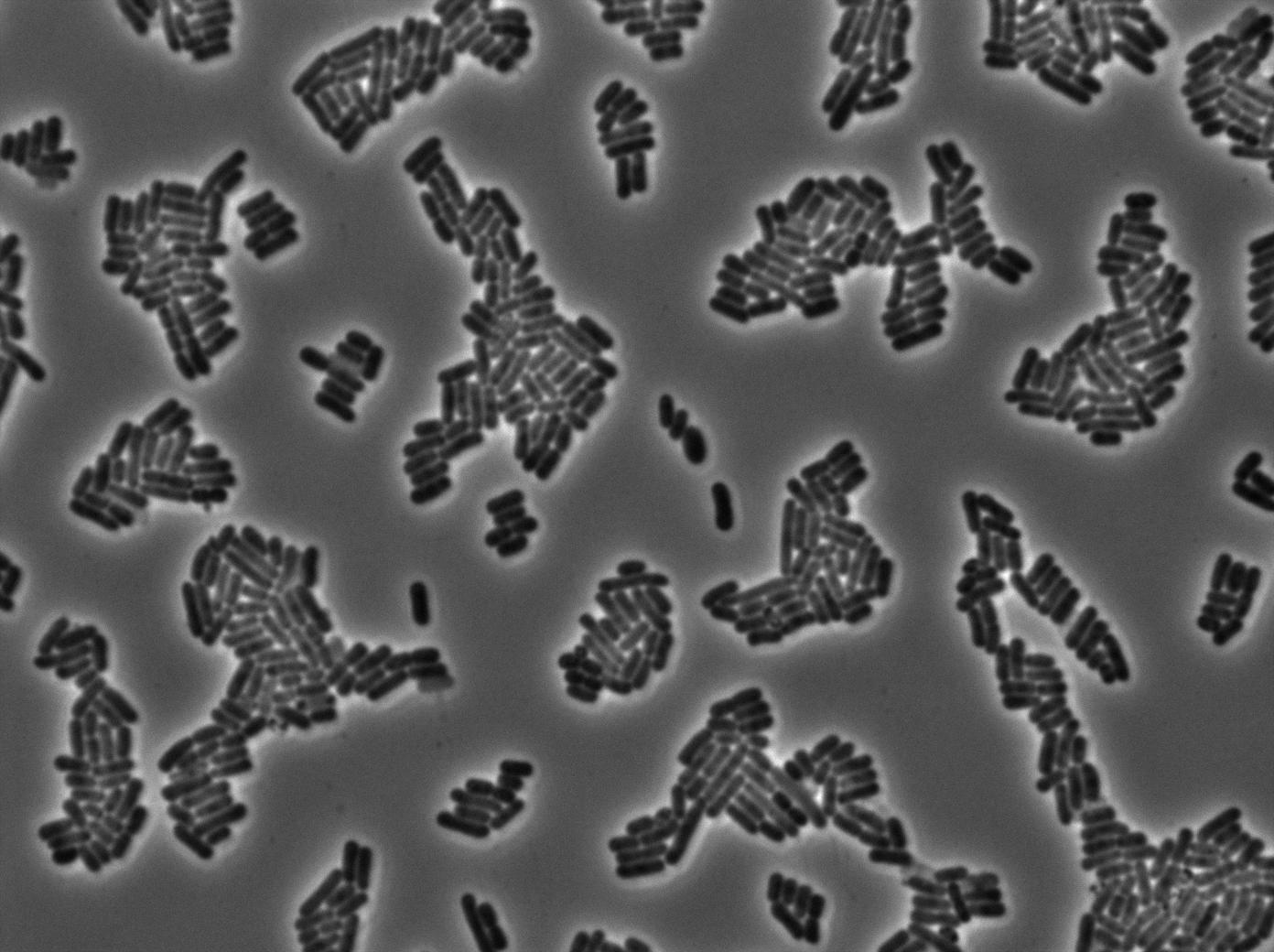
We characterized T7 autoloop in E.coli, when hosted in the plasmid pSB1C3 (K606036). The idea was to see if the system was leaky inside a synthetic biology plasmid, that is to say, a plasmid holding 4 terminators before any construct.
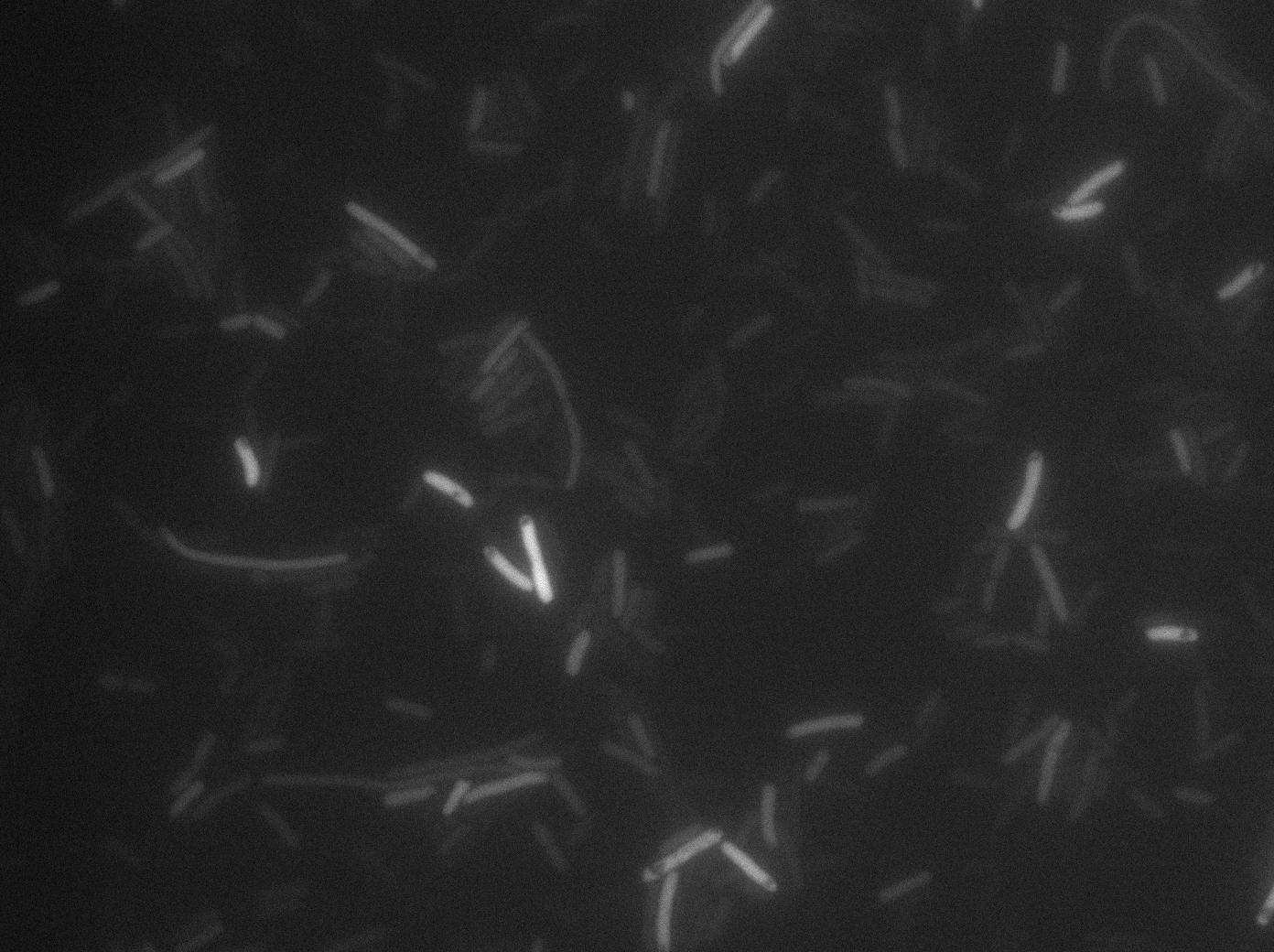
We found out there is a small leakage with the T7 emitter RFP, the cells in which the positise feedback loop is activated stop dividing and glow with a very strong signal.
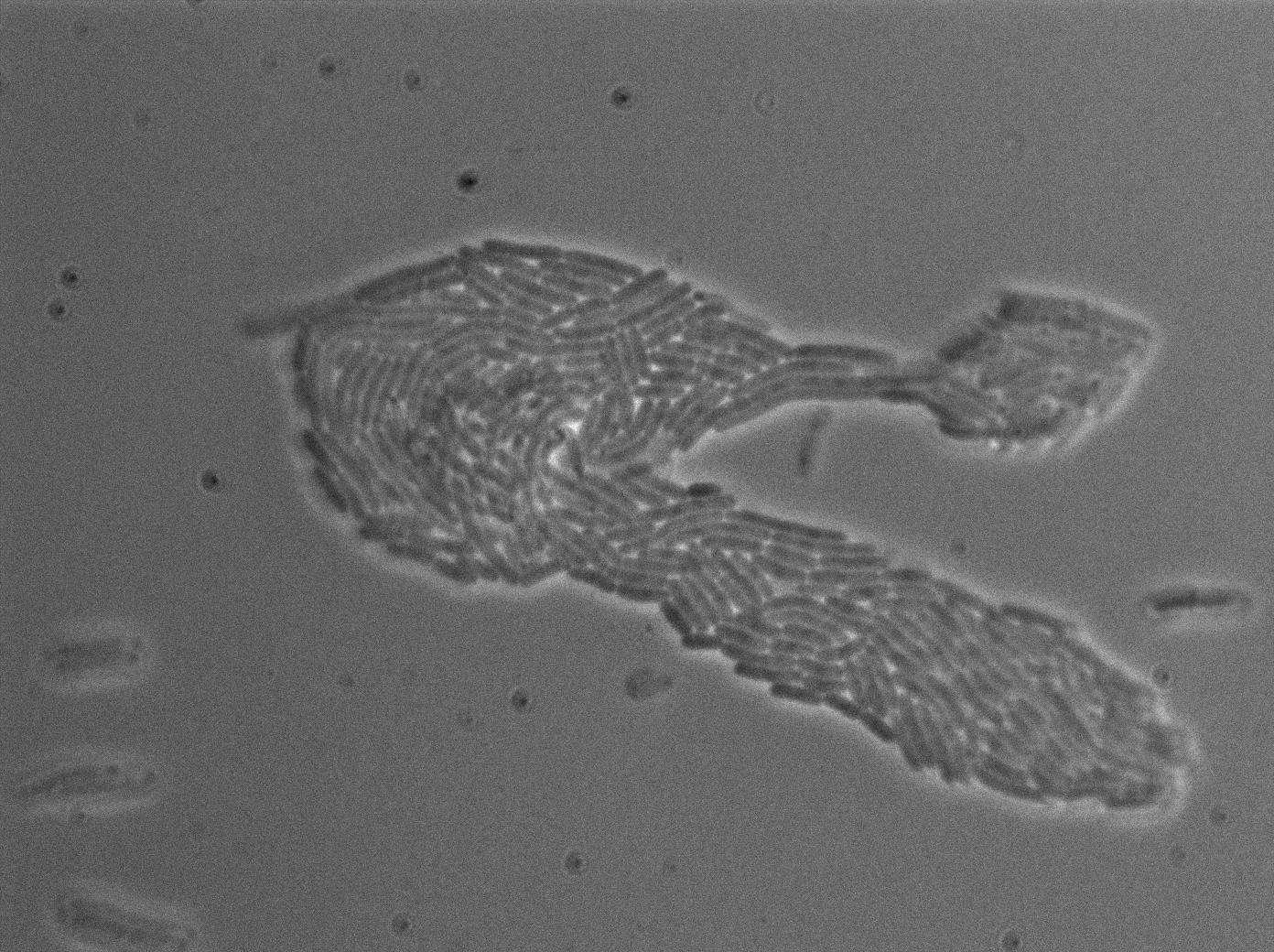
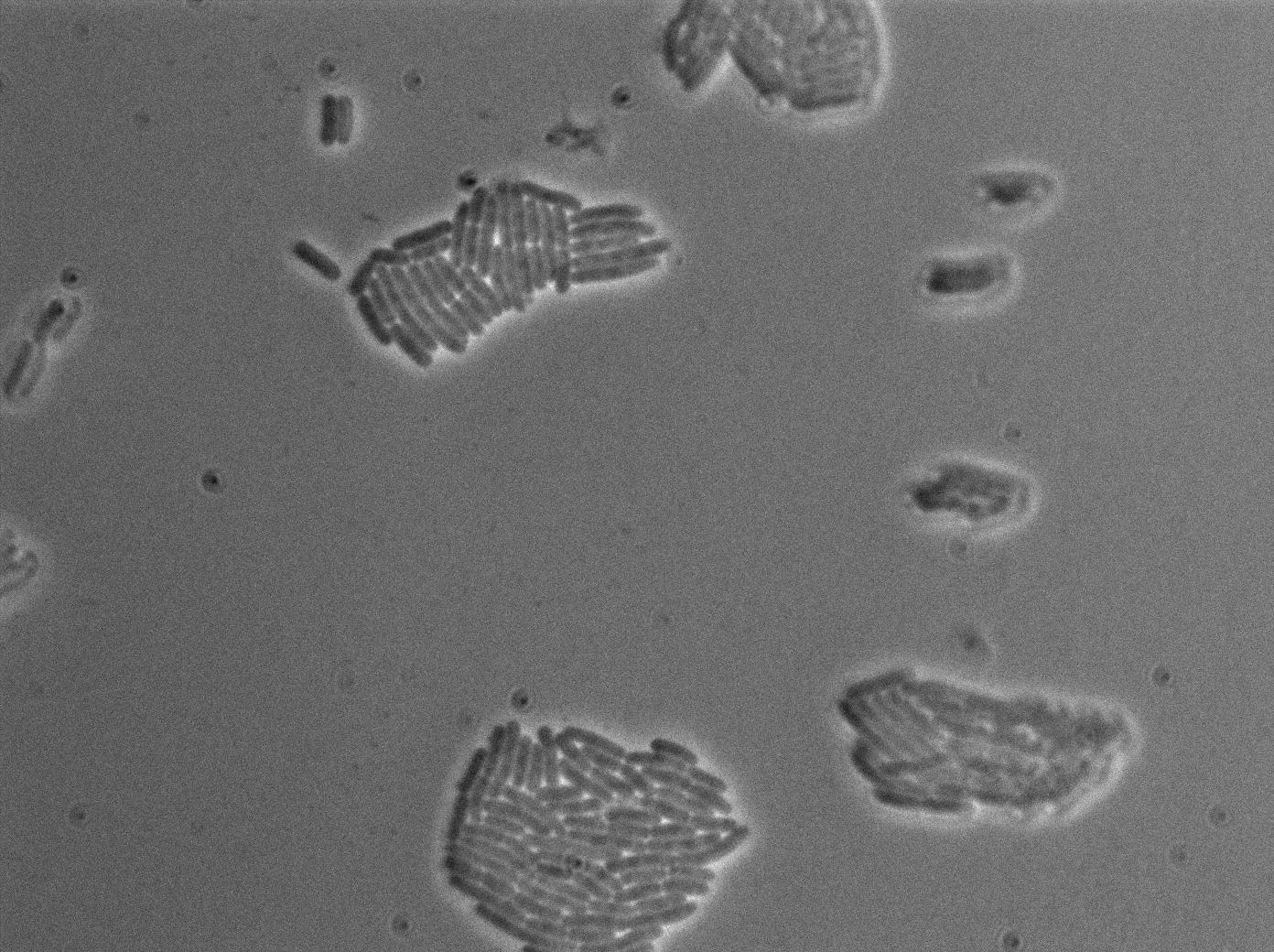
The first pictures show the RFP construct has been well done since some cells are glowing with RFP fluorescence. This also shows the system ins not as leaky as we expected for. Indeed, the promoter regulating RFP expression is 'pVeg' which is a constitutive promoter. Finally, the RFP system is working very well because when leak occurs, cells glow highly.
These pictures show that GFP system gives more disparity than the RFP construct. Moreover, the absence of double terminator could explain the leakiness of our system. However, we expected this cells to glow in RFP but they did not. This shows that the full construct has failed.
The last pictures show less disparity that the previous one. This shows that double terminator actually proceeds on the phenotype. We also can notice that the cells glow in red a bit. However, we can not say if it is an artifact or if the system is actually well constructed with the terminator and RFP construct.
Future
In order to get a better characterization of these BioBricks and tweak their parameters, we have already built the microfluidic device.
Afterwards, we want to clone the LacI expression system (K606054) into the emitter cell (K606039) in order to get the T7 controlled by LacI and thus to improve the expression of T7 polymerase.
Finally, we plan to insert again these BioBricks in a replicative vector like pHM3 or an integrative vector (like K090403) for B.subtilis in order to test the communication and the transfer of T7 polymerase through the nanotubes and so characterize them.
 "
"