Team:DTU-Denmark/Project
From 2011.igem.org
Project
Contents |
Tuning regulation with a non-coding RNA trap
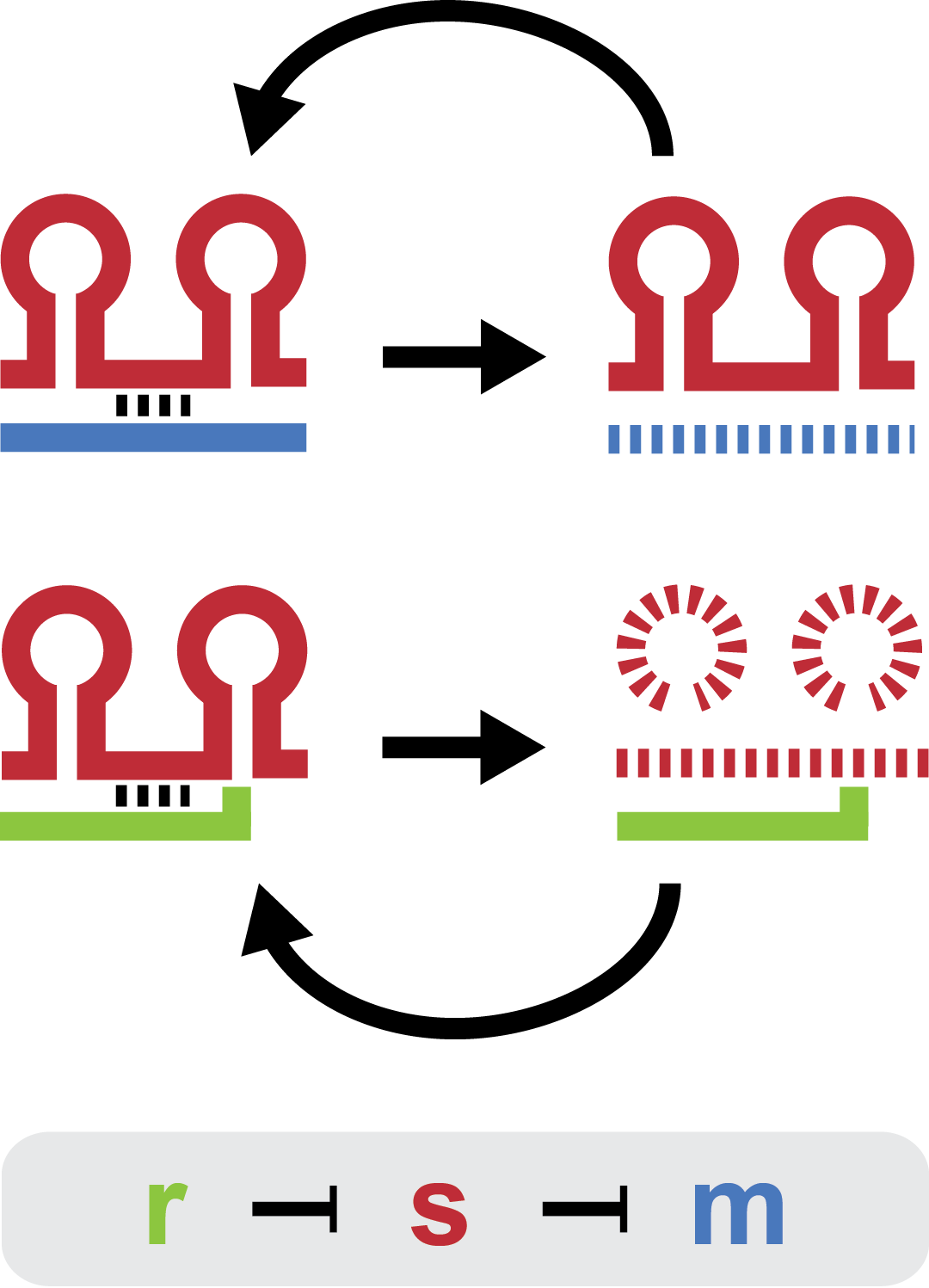
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. One such application could be the optimization and fine-tuning of synthetic biological circuits, which is currently a cumbersome process of trial and error. We have investigated a novel type of RNA regulation, where the inhibition caused by a small regulatory RNA is relieved by another RNA called trap-RNA. The system displays a large dynamic range and can uniquely target and repress any gene of interest providing unprecedented flexibility. We suspect that any level of repression is achievable by simply altering the sequences of the involved RNAs. Multiple such systems can coexist without interfering and are thus compatible with more complex designs. Furthermore the trap-RNA can be fused to any transcript in effect allowing any gene to act as an activator.
Overview
Our project is a proof of concept project, showing that the E. coli sRNA sroB and the intergenic sRNA in the chbBCARG gene can be used to control gene expression by targeting the ybfM Shine-Delgarno, and that these sRNAs can be rationally designed to target other Shine-Delgarnos.
The experimental part of the project can be broken into 3 distinct parts, which combined form the complete project:
- Construction of plasmids
- Strain construction
- Improving the araBAD promoter
Construction of plasmids necessary for testing our system involves taking the native system from E. coli, as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system.
Strain construction involves deleting the original genes from the chromosome of a E. coli W3110 strain. Since we use the original genes, these need to be deleted from the chromosome to prevent them from interfering with our measurements.
Improving the araBAD promoter entails expanding the dynamic range of this promoter by modifying the -10 and -35 sequence of the promoter, as well as randomly changing the nucleotide sequence around and in between these sequences. Since the araBAD promoter is used in our project improving this promoter could lead to even finer control of our system.
Plasmids
There are three elements in the natural trap RNA system that regulate chitobiose metabolism in E. coli: (1) chitoporin chiP (alias ybfM) that facilitates uptake of chitosugars from the medium into the cell; (2) sRNA chiX (alias sroB, micM) which post-transcriptionally regulates chiP; and (3) chiXR sRNA which is transcribed from intergenic region in chbBCARG operon and regulates chiX sRNA. ChiX sRNA regulates chiP expression through binding to the Shine-Dalgarno sequence on the chiP mRNA and thus inhibits recruitment of 30S ribosome subunit and translation cannot occur. Only when chitobiose is present in the medium can the chiXR be transcribed and subsequently its transcript can bind to chiX sRNA, releasing chiP repression.
In order to quantitatively describe the natural trap RNA system following three plasmids were constructed:
- PchiP-lacZ plasmid
- To mimic and test repression of the chiP gene its upstream region of 599 bp was fused to reporter genes lacZ and GFP and inserted on the pSLD39 plasmid. This constructs were controlled by constitutive promoter. LacZ codes for $\beta$-galactosidase and its activity was tested using $\beta$-Gal assay while GFP activity was tested using Biolektor.
- Ptet-chiX plasmid
- ChiX, which is 84 bp long, was inserted into pSB4K plasmid together with regulatory element from tet operon. Expression from Ptet promoter was induced using anhydrotetracycline.
- PBAD-chiXR plasmid
- Introgenic region from chbBCARG operon 284 bp long was inserted into pBAD18 plasmid. Expression from that plasmid was controlled by regulatory element from ara operon with arabinose as inducer.
Strain
The strain used in testing our system is based on the E. coli strain W3110, a wild-type strain of Escherichia coli K-12 \cite{Hayashi2006} with the lacZYA, chbBCARG operons and chiP (alias ybfM), chiX (alias sroB, micM) genes deleted from the chromosome. The three latter code for utilization of chitobiose (a sugar), chitobiose permease and the regulation of the chitobiose permease respectively.
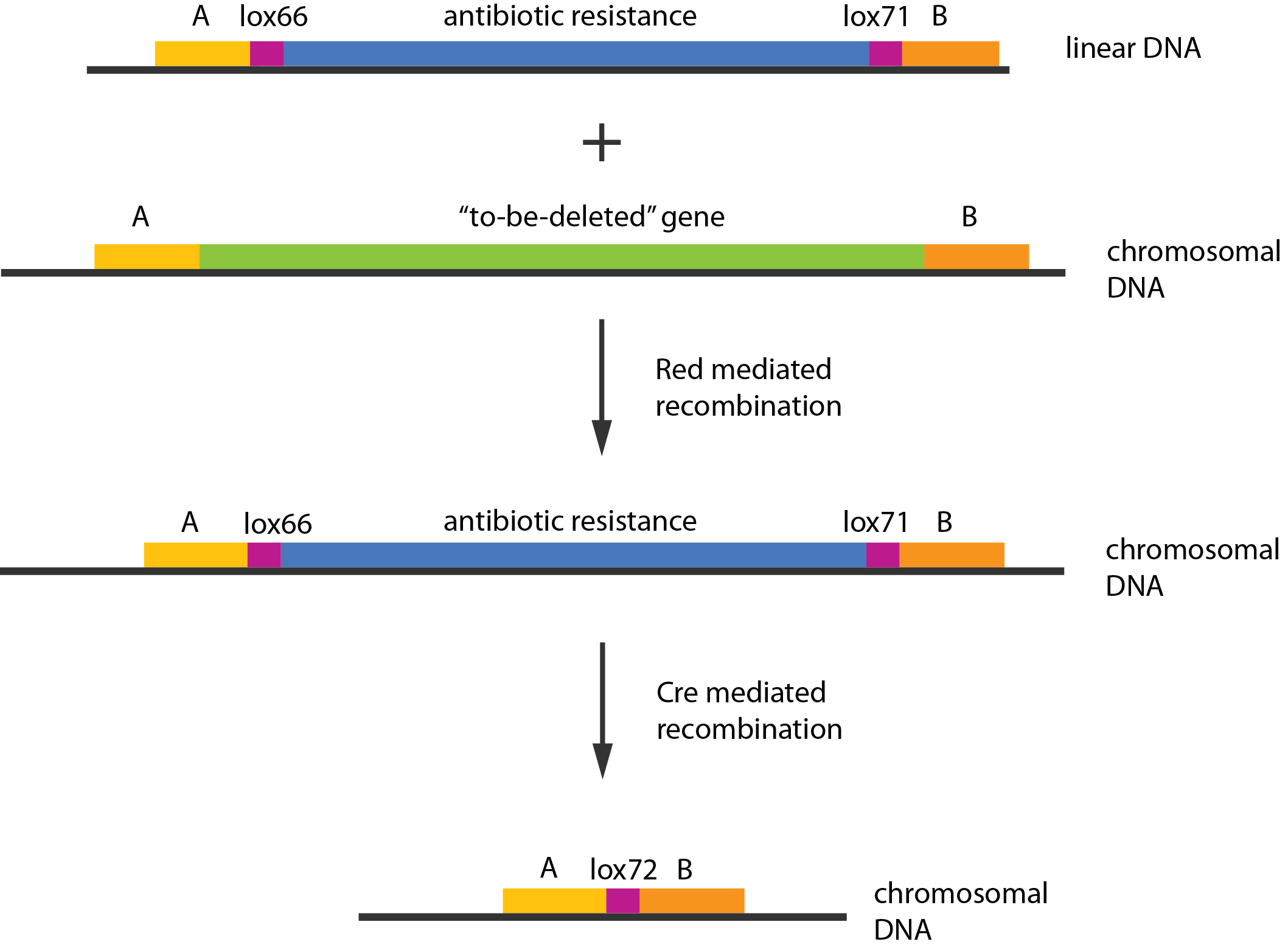
A procedure known as redswap \cite{Datsenko2000} was used to delete these genes. Schematically the procedure works in the following manner:
- Transform competent cells with Red and cre helper plasmids.
- Make the cells competent and induce the Red recombinase.
- Transform with linear piece of DNA.
- Select for gain of resistance.
- Move to medium inducing the Cre recombinase.
- Screen for loss of resistance.
- Verify the construct.
- Repeat step 2-7 as needed.
- Cure the helper plasmids.
The Red helper plasmids encodes the $\lambda{}$ phage Red recombination system. This system allows recombination events between sequences with around 40 nucleotides of homology. The cre helper plasmid encodes the Cre recombinase. This recombinase recognises 34bp loxP sequences and promotes recombination between them.
An example of a linear piece of DNA that could be used in redswap, as well as the simplified representation of the procedure, can be seen in figure \ref{linDNA}.
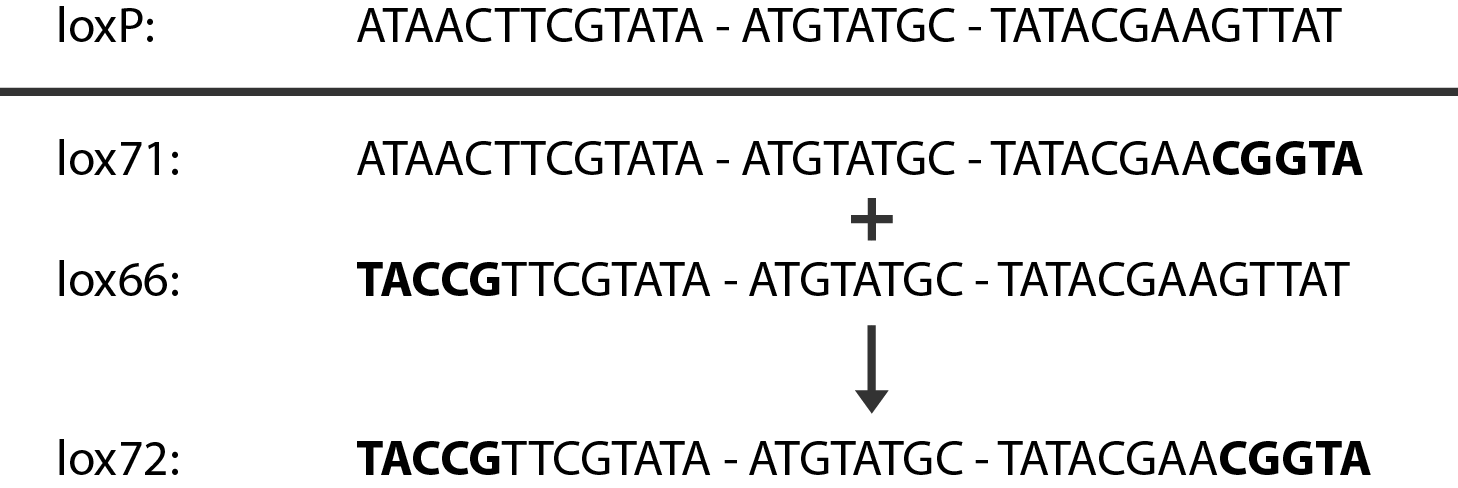
In case of two or more subsequent gene deletions using redswap a problem of recombination between an old loxP site and newly introduced ones arises. Remember, that after each round of redswap one loxP site is left in the genome. As a result recombination using Cre recombinase in second or next redswap might not occur between loxP sites flanking a resistance gene. To circumvent this problem two mutated loxP sequences are used - lox66 and lox71 \cite{Lambert2007}. In spite of the mutations these lox sites are recognized by Cre which can remove the intervening region. Finally, from lox66 and lox71 a new site is formed, lox72, which has a reduced affinity to Cre recombinase. Thus, more than one gene deletions can be subsequently conducted.
 "
"