Team:EPF-Lausanne/Tools/Gibson assembly
From 2011.igem.org
Gibson assembly
Gibson assembly is a useful DNA ligation method that offers an alternative to the classical approach of using restriction enzymes. The procedure was was developed in 2009 by Gibson et al [http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html] and wsa introduced to the iGEM competition by the Cambridge 2010 team.
Requirements
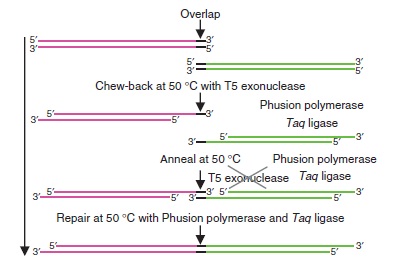
In order to perform Gibson assembly on two DNA sequences, these sequences need to overlap by at least 20bp at the edge. You can easily add these overlaps by extension PCR
Principle
Gibson assembly relies on the activity of 3 different enzymes:
- T5 exonuclease
- Phusion polymerase
- Taq ligase
Combined together, these 3 enzymes allow overlapping DNA sequences (sequences A and B) to be ligated together in a single and isothermal reaction. First, the T5 exonuclease chews up bases at the 5' end on each strand, creating single-strands on the edges. Due to the sequence overlap, the single strand from A will anneal with the single strand from B. Then, the Phusion polymerase extend the DNA sequence from the 3' of the AB overlap. Finally, when there all the gaps have been filled by the polymerase, the Taq ligase ties up the A and B fragments.
 "
"