Team:UNAM-Genomics Mexico/Project/RhizobialKit
From 2011.igem.org
General Overview
Abstract
Expanding our ability to manipulate and engineer biological parts in new organisms is an ever present goal of synthetic biology. With this idea in mind we set out to develop a kit of biological tools to enable working with BioBricks in Rhizobial species. We developed the tools in this kit with our model chassis being Rhizobium etli, a member of the Rhizobiaceae family of nitrogen fixating bacteria, that establishes a symbiotic relationship with legumes within the root nodules.
Contents |
Rhizobial Kit Description
The Rhizobial Kit will set the ground for the use of Rhizobial species, especially R. etli, as organisms available for the design and construction of novel synthetic biological machines. The kit will be composed of parts with distinct molecular functions that will increase the repertoire of possible design and engineering approaches in Rhizobial species.
The parts that we will develop are the following:
- The nifH promoter. This promoter sequence comes from the promoter region of the nifHa gene in R. etli. This promoter region contains a σ54-dependent promoter located between position -24 and -12 relative to the transcription start site, as well as a binding site for NifA, a positive transcriptional regulator. The binding of the NifA activator to the promoter region promotes transcription in conditions of low oxygen concentration, and the -24/-12 conserved bases have been implicated in the context of nitrogen fixation and regulation (Beat Thöny, Hauke Hennecke, 1989; Valderrama, B., et. al., 1996).
- A re-characterization of the Anderson Promoter Collection in R. etli. These constitutive promoters are already characterized in E. coli [http://partsregistry.org/Promoters/Catalog/Anderson], and our goal is to investigate the strength of these promoters in R. etli as the chasis. This will hopefully give a spectrum of promoters with different strengths that will allow the fine-tuning of the expression of devices in this model.
- We developed a standardized plasmid that is able to integrate BioBrick constructions into Rhizobial species after conjugation with transformed E. coli cells. This plasmid is a modification of the pBBR1MCS-5 plasmid, which is a broad-host-range (bhr) vector designed to assist genetic anaylsis in prokaryotes, specifically for Gram- bacteria that are naturally CmR (chloramphenicol resistant). It is used to transfer genes from E. coli to R. etli by first transforming E. coli S17 competent cells with this vector containing an RFP reporter and later performing a conjugation between the transformed E. coli cells and R. etli CFN42.
- A repC “plasmid replication device” as a standard part is included. Its purpose is to render a plasmid capable of repC-dependent replication in R. etli. The repC region contains two genes: one encoding the initiator protein RepC (46.8 kDa) and other, an antisense RNA (67 nt). repC is the minimal region required for stable replication of a member of the repC plasmid family.
Rhizobial Kit Details
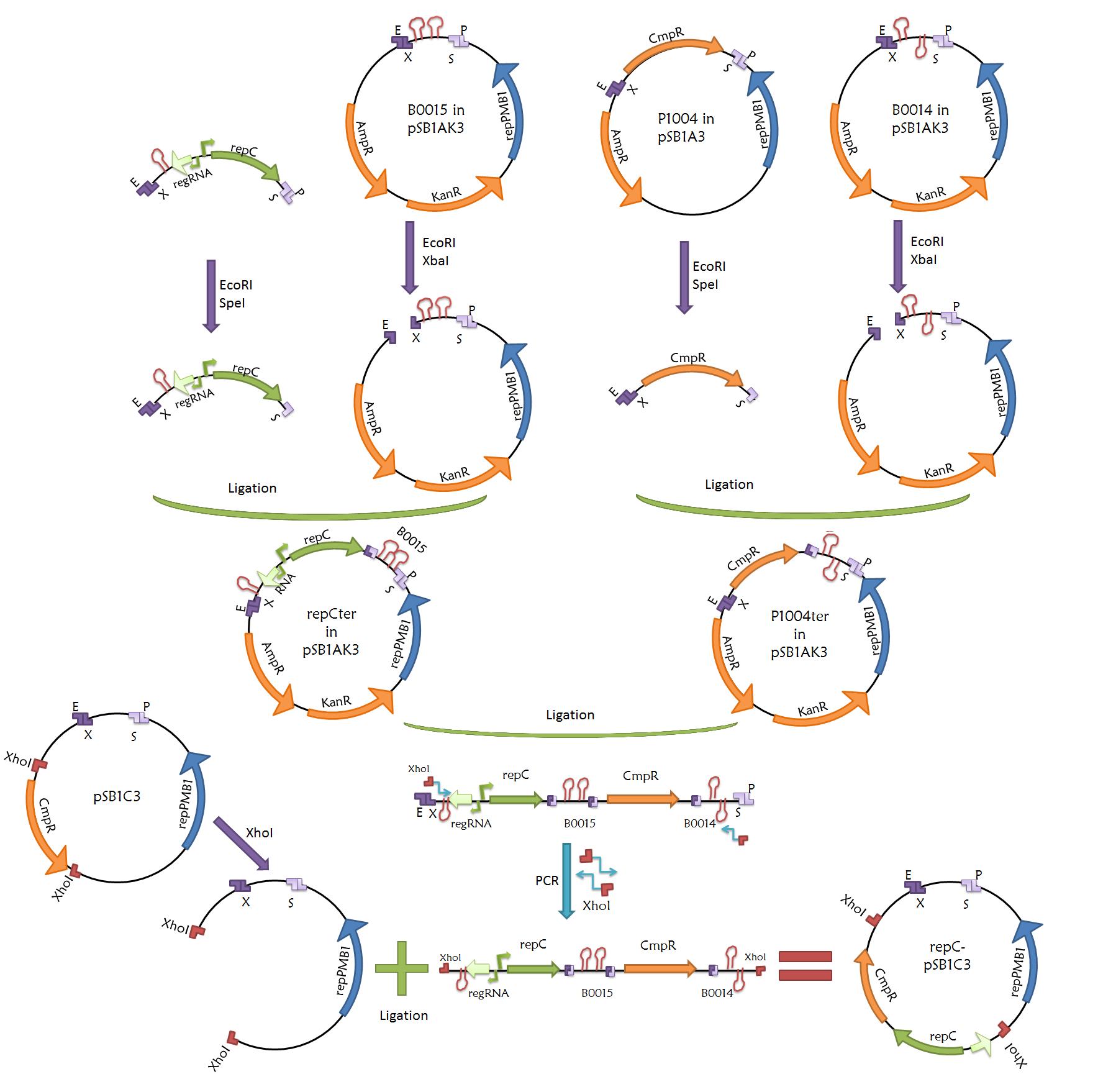
- pBBR1MCS-5 derived plasmid. This is a general scheme showing the methodology used to standardize the plasmid, the strategy is based on inserting a segment of the pSB1T3 which contains the prefix, suffix and terminators into the pBBR1MCS-5
- Plasmid with RepC. This is the final construction of the second plasmid
Justification
The main goal of our project is to produce molecular hydrogen as an energy transporter in R. etli cells in a auto-sustainable fashion. While pursuing this objective we faced the necessity of generating basic standard tools to systematically characterize the components of our system. It became clear that expanding the repertoire of standard parts available for their use in our model system, R. etli, was not only beneficial to our causes, but touched on a basic purpose of synthetic biology: broadening the range of organisms in which rationalized engineering approaches help develop and characterize biological devices. Our model chassis and related species stand out for their nitrogen fixation capacity, and the transcriptional regulation dependent on anaerobic or hypoxic conditions. Thus, with the development of these bioengineering tools, we are extending the reach of synthetic biology to new crowds, questions, and horizons.
References
- Brenda Valderrama, Araceli Dávalos, Lourdes Girard, Enrique Morett. 1996. Regulatory proteins and cis-acting elements involved in the transcriptional control of Rhizobium etli reiterated NifH genes. Journal of Bacteorology, 178, 3119-3126.
- Beat Thöny, Hauke Hennecke. 1989. The -24/-12 promoter comes of age. Microbiology Reviews 63, 341-358.
- Michael E. Kovach, Philip H. Elzer, D. Steven Hill, Gregory T. Robertson, Michael A. Farris,R. Martin Roop, Kenneth M. Peterson (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175-176
 "
"