Team:Edinburgh/Cell Display
From 2011.igem.org
| Line 57: | Line 57: | ||
This will not be a problem in the JM109 lab strain, which lacks an important <span class="hardword" id="recombination">recombination</span> enzyme. As for the use of this technology in industry, it will be possible to overcome this problem simply by synthesising coding sequences with as many altered (but <span class="hardword" id="synonymouscodon">synonymous</span>) codons as possible. We have written a software tool for designing such sequences... see the [[Team:Edinburgh/Genetic instability|genetic instability]] page. | This will not be a problem in the JM109 lab strain, which lacks an important <span class="hardword" id="recombination">recombination</span> enzyme. As for the use of this technology in industry, it will be possible to overcome this problem simply by synthesising coding sequences with as many altered (but <span class="hardword" id="synonymouscodon">synonymous</span>) codons as possible. We have written a software tool for designing such sequences... see the [[Team:Edinburgh/Genetic instability|genetic instability]] page. | ||
| - | == | + | ==Proof of concept== |
| - | + | A working proof of concept could involve simply displaying the Yellow Fluorescent Protein on INP. Indeed, something similar was achieved by [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2009.01724.x/abstract Li ''et al'' (2009)]. | |
| - | + | ||
| - | + | ||
==References== | ==References== | ||
| + | |||
| + | * Li Q, Yu Z, Shao X, He J, Li L (2009) [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2009.01724.x/abstract Improved phosphate biosorption by bacterial surface display of phosphate-binding protein utilizing ice nucleation protein]. ''FEMS Microbiology Letters'' '''299'''(1): 44-52 (doi: 10.1111/j.1574-6968.2009.01724.x). | ||
* Van Bloois E, Winter RT, Kolmar H, Fraaije MW (2011) [http://www.sciencedirect.com/science/article/pii/S016777991000199X Decorating microbes: surface display of proteins on ''Escherichia coli'']. ''Trends in Biotechnology'' '''29'''(2): 79-86 (doi: 10.1016/j.tibtech.2010.11.003). | * Van Bloois E, Winter RT, Kolmar H, Fraaije MW (2011) [http://www.sciencedirect.com/science/article/pii/S016777991000199X Decorating microbes: surface display of proteins on ''Escherichia coli'']. ''Trends in Biotechnology'' '''29'''(2): 79-86 (doi: 10.1016/j.tibtech.2010.11.003). | ||
| - | + | ||
</div> <!-- /main_body--> | </div> <!-- /main_body--> | ||
<html></div> <!-- /mids --></html> | <html></div> <!-- /mids --></html> | ||
Revision as of 15:38, 12 August 2011
An obvious type of bioreactor is an E. coli cell that displays the desired proteins on its outer membrane. This type of display is called cell surface display.
This works by fusing the proteins of interest to carrier proteins which are naturally found on the outer membrane.
Contents |
Outline
Berkeley 2009 tried several different carrier proteins with several different passenger enzymes, and had success in many areas. However, when they tried attaching cellulases, they weren't so successful - of the two quantified cellulases, one worked just as well without the carrier (Cel5b) and the other didn't work (Cel9a, as compared to negative control).
We will try a different carrier. <partinfo>BBa_K265008</partinfo> by UC Davis 2009 is a synthetic, codon-optimised sequence, based on [http://www.ncbi.nlm.nih.gov/nuccore/AF013159 GenBank AF013159] and coding for the first 211 and last 97 amino acids of ice nucleation protein (INP, normally coded by the inaK gene) from the bacterium Pseudomonas syringae. It seems promising as a carrier of enzymes. Fusions are carried out at the INP C terminal.
[http://www.sciencedirect.com/science/article/pii/S016777991000199X Van Bloois et al (2011)] speak highly of INP, and claim that it can be displayed at a copy number of around 100,000 copies per cell without affecting viability.
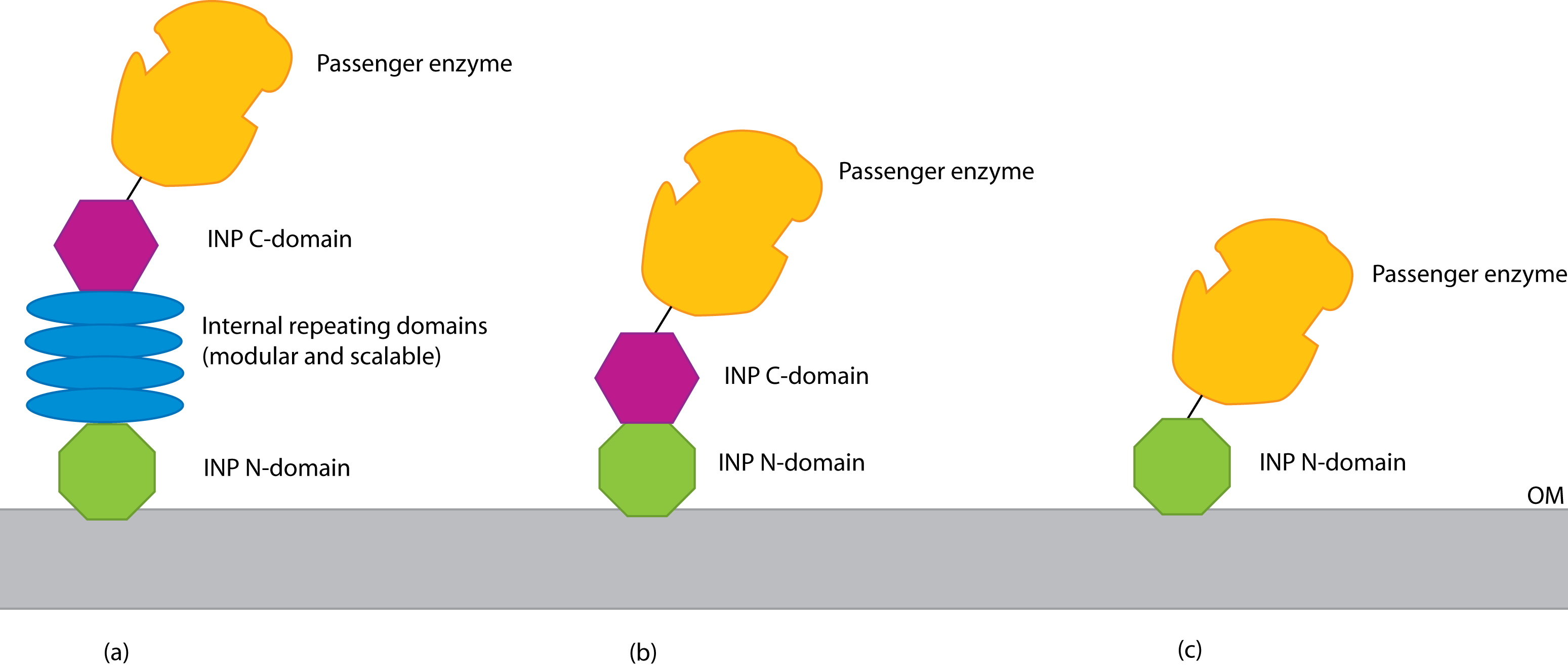
INP has major domains at its N and C terminals, as well as a number of internal repeating domains. There seem to be three strategies for using INP (see figure):
- Use the entire INP protein; fuse at the C terminal
- Delete the INP internal domains; fuse at the C terminal
- Delete all of INP except the N domain; fuse at the new C terminal
<partinfo>BBa_K265008</partinfo> should be suitable for the 2nd strategy.
Linkers
It may be desirable to create linkers between the carrier and the protein of interest. BioSandwich could be ideal for this.
Example system
A complete 3 cellulase system would contain:
- Promoter -- RBS -- <partinfo>BBa_K265008</partinfo> -- Linker? -- <partinfo>BBa_K392006</partinfo>
- Promoter -- RBS -- <partinfo>BBa_K265008</partinfo> -- Linker? -- <partinfo>BBa_K392007</partinfo>
- Promoter -- RBS -- <partinfo>BBa_K265008</partinfo> -- Linker? -- <partinfo>BBa_K392008</partinfo>
An alternative: protein chains
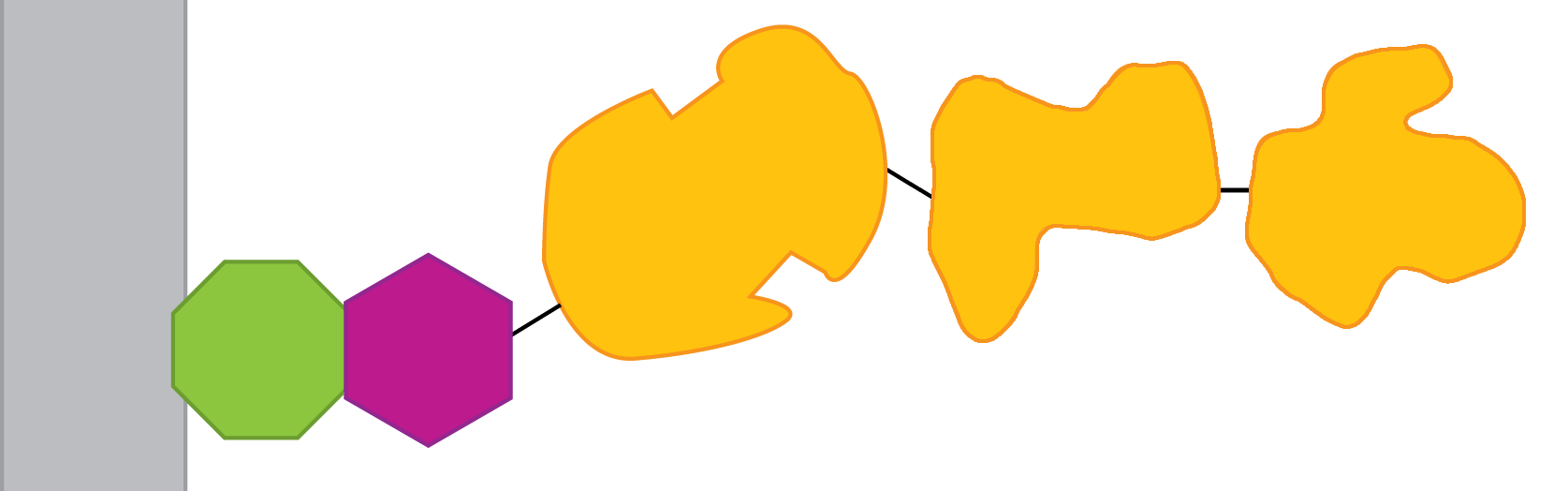
Instead of making three different fusions, it might be possible to make one fusion that had all three cellulase enzymes linked together:
- Promoter -- RBS -- <partinfo>BBa_K265008</partinfo> -- <partinfo>BBa_K392006</partinfo> -- <partinfo>BBa_K392007</partinfo> -- <partinfo>BBa_K392008</partinfo>
If this works, it would be interesting to optimise the order in which the proteins appear.
However, the size of the chain exceeds the size of proteins that have been successfully displayed on INP. It might be possible to remove unnecessary domains, e.g. cellulose-binding domains.
Genetic instability
In order to display several different proteins on one bacterium using the first strategy, it will be necessary to have several copies of the INP gene fused to different enzymes. The presence of repeated sequences on a plasmid can lead to genetic instability.
This will not be a problem in the JM109 lab strain, which lacks an important recombination enzyme. As for the use of this technology in industry, it will be possible to overcome this problem simply by synthesising coding sequences with as many altered (but synonymous) codons as possible. We have written a software tool for designing such sequences... see the genetic instability page.
Proof of concept
A working proof of concept could involve simply displaying the Yellow Fluorescent Protein on INP. Indeed, something similar was achieved by [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2009.01724.x/abstract Li et al (2009)].
References
- Li Q, Yu Z, Shao X, He J, Li L (2009) [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2009.01724.x/abstract Improved phosphate biosorption by bacterial surface display of phosphate-binding protein utilizing ice nucleation protein]. FEMS Microbiology Letters 299(1): 44-52 (doi: 10.1111/j.1574-6968.2009.01724.x).
- Van Bloois E, Winter RT, Kolmar H, Fraaije MW (2011) [http://www.sciencedirect.com/science/article/pii/S016777991000199X Decorating microbes: surface display of proteins on Escherichia coli]. Trends in Biotechnology 29(2): 79-86 (doi: 10.1016/j.tibtech.2010.11.003).
 "
"