Team:Paris Bettencourt/Modeling/T7 diffusion
From 2011.igem.org
(→Design) |
(→Design) |
||
| Line 27: | Line 27: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | <p>The reporter for the emitter gene construct ( | + | <p>The reporter for the emitter gene construct (<em>RFP</em>) is modeled by the following equations:</p> |
<img src='https://static.igem.org/mediawiki/2011/9/95/RFP_equations.png' style='width:100%;'> | <img src='https://static.igem.org/mediawiki/2011/9/95/RFP_equations.png' style='width:100%;'> | ||
<br> | <br> | ||
| Line 34: | Line 34: | ||
<p>The receiver and amplification gene construct is modeled by the following equations:</p> | <p>The receiver and amplification gene construct is modeled by the following equations:</p> | ||
<img src='https://static.igem.org/mediawiki/2011/f/f2/T7%27%27_equations.png' style='width:100%;'> | <img src='https://static.igem.org/mediawiki/2011/f/f2/T7%27%27_equations.png' style='width:100%;'> | ||
| + | <br> | ||
| + | <br> | ||
| + | <p>The reporter for the receiver and amplification gene construct (<em>GFP</em>) is modeled by the following equations:</p> | ||
| + | <img src='https://static.igem.org/mediawiki/2011/2/2f/GFP_equations.png' style='width:100%;'> | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 08:25, 12 August 2011

Contents |
Model for T7 diffusion
Design
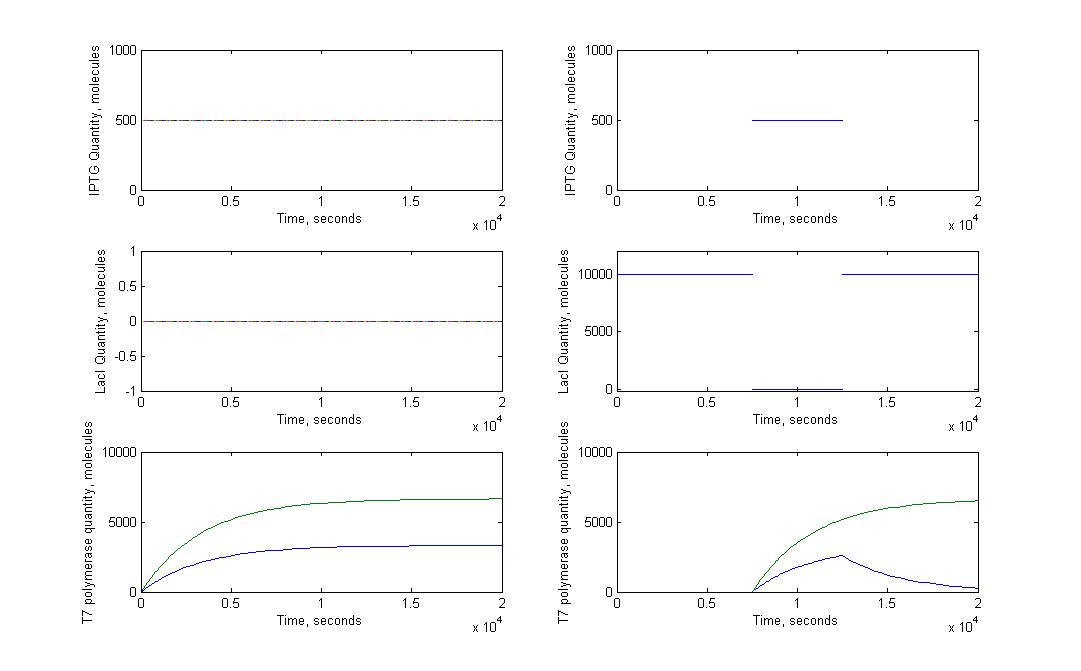
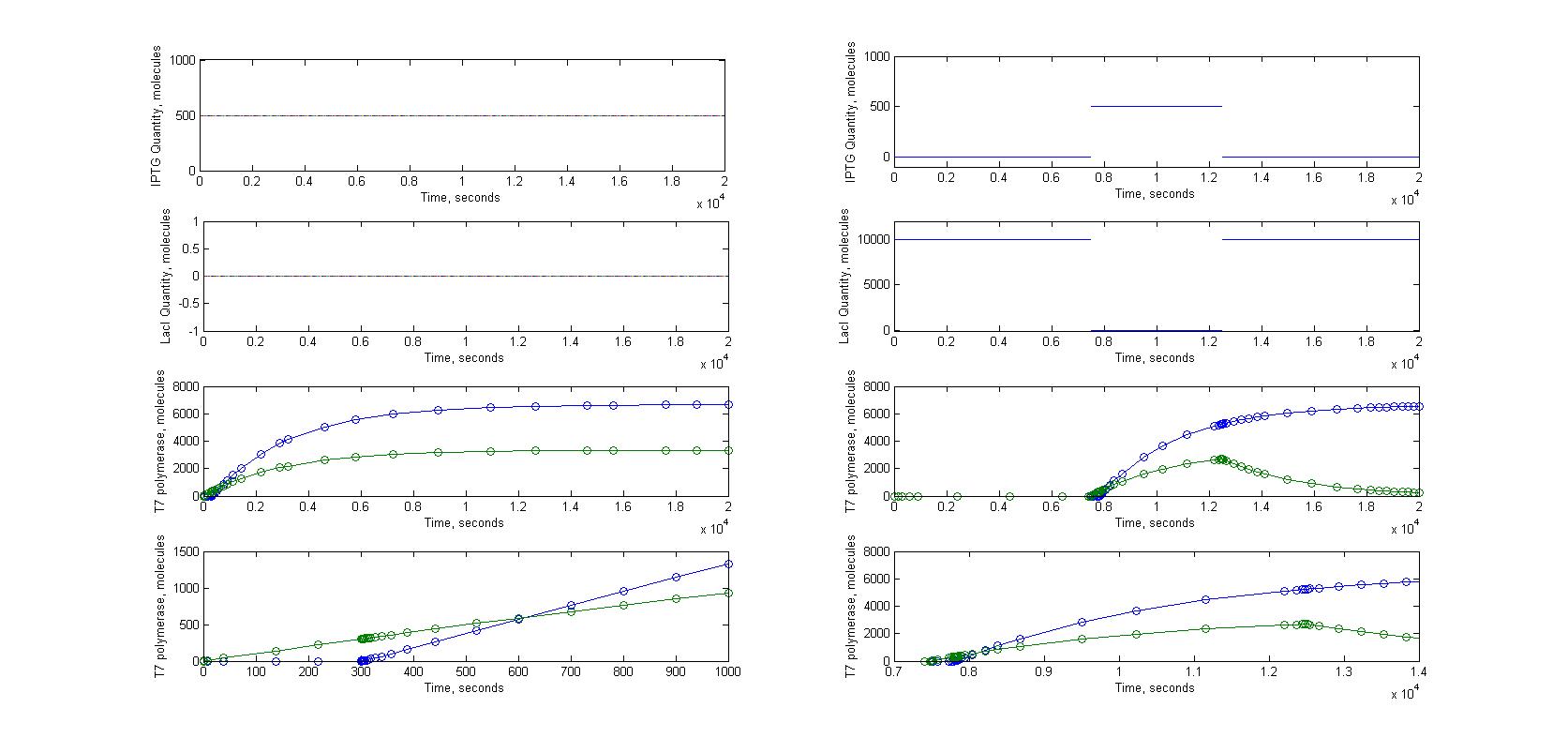
The T7 diffusion design is our first original construct. In this design, the T7 RNA polymerase acts both as the signal transmited and as the amplifier (auto-amplification). T7 RNA polymerase is produced in the emitter cell, then diffuses through the nanotubes and arrives in the receiver cell. In this cell, the T7 RNA polymerase activates on a pT7 promoter. Behind this promoter, we put a gene coding for the same T7 RNA polymerase (amplifier) and for GFP (reporter).
This construction was put in two different settings. One is what we just described, where the emitting gene network is in one cell and the receiving gene network is in another. In the other construction, everything is in one cell. We use the second construct as a control to really see the impact of the cell-to-cell communication on the behaviour of the cells.
We ran our models for those two configurations. We used a steady flow of signaling molecules in the receiver cell for the "one emitting cell - one receiving cell" construction. You can find our justifications about this assumption here.
Model
We use the ''LacI'' as a repressor for the emitter gene construct. LacI repression can be cancelled by ''IPTG''. This way we can induce production of RFP and ''T7''' by puttig ''IPTG'' on the cells.


Inactivated LacI can not repress the pLAC promoter anymore. Note that we consider that the reaction between IPTG and LacI fires without any delay. This assumption is justified by the fact that this reaction is much faster than any other in our gene network.
The emitter gene construct is modeled by the following equations:

The reporter for the emitter gene construct (RFP) is modeled by the following equations:

The receiver and amplification gene construct is modeled by the following equations:

The reporter for the receiver and amplification gene construct (GFP) is modeled by the following equations:

Parameters
This design relies on the T7 RNA polymerase (which is noted T7) both as the signaling molecule going through the nanotubes and as the auto-amplification system when it acts on the pT7 promoter. In our equations however, we chose to distinguish these functions.
- T7' represents the signaling T7 RNA polymerase.
- T7'' represents is the auto-amplifying molecule.
 INSERT JAVASCRIPT TO HIDE/SHOW
INSERT JAVASCRIPT TO HIDE/SHOW
 INSERT JAVASCRIPT TO HIDE/SHOW
INSERT JAVASCRIPT TO HIDE/SHOW
The parameters used in this model are:
| Parameter | Description | Value | Unit | Reference |
|---|---|---|---|---|
 |
Active LacI concentration (LacI which is not inactivated by IPTG) | NA | molecules per cell |
Notation convention |
 |
IPTG concentration | NA | molecules per cell |
Notation convention |
 |
Inactived LacI concentration | NA | molecules per cell |
Notation convention |
 |
Total LacI concentration | TBD | molecules per cell |
Steady state for equation |
 |
T7 RNA polymerase (emitter, T7') concentration | NA | molecules per cell |
Notation convention |
 |
mRNA associated with T7' concentration | NA | molecules per cell |
Notation convention |
 |
T7 RNA polymerase (auto-amplification, T7'') concentration | NA | molecules per cell |
Notation convention |
 |
mRNA associated with T7'' concentration | NA | molecules per cell |
Notation convention |
 |
Maximal production rate of pVeg promoter (constitutive) | ??? | molecules.s-1 or pops |
Estimated |
 |
Maximal production rate of pLac promoter | 0.02 | molecules.s-1 or pops |
Estimated |
 |
Maximal production rate of pT7 promoter | 0.02 | molecules.s-1 or pops |
Estimated |
 |
Dissociation constant for IPTG to LacI | 1200 | molecules per cell |
Aberdeen 2009 wiki |
 |
Dissociation constant for LacI to LacO (pLac) | 700 | molecules per cell |
Aberdeen 2009 wiki |
 |
Dissociation constant for T7 RNA polymerase to pT7 | 3 | molecules per cell |
Estimated ADD EXPLANATION |
 |
Translation rate of proteins | 1 | s-1 | Estimated ADD EXPLANATION |
 |
Dilution rate in exponential phase | 2.88x10-4 | s-1 | Calculated with a 40 min generation time. See explanation |
 |
Degradation rate of mRNA | 2.88x10-3 | s-1 | Uri Alon (To Be Confirmed) |
 "
"