Team:EPF-Lausanne/Our Project/Reporter Systems/ptet
From 2011.igem.org
(→Ptet Characterization) |
(→Ptet Characterization) |
||
| Line 6: | Line 6: | ||
Since we are using the Ptet promoter in combination with TetR in our reporter systems, it was useful for us to characterize the Ptet promoter only. This characterization was done with the [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/Reporter_Systems/plasmid#First_reporter_-_J61002_Ptet-RFP J61002 Ptet-RFP] plasmid. | Since we are using the Ptet promoter in combination with TetR in our reporter systems, it was useful for us to characterize the Ptet promoter only. This characterization was done with the [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/Reporter_Systems/plasmid#First_reporter_-_J61002_Ptet-RFP J61002 Ptet-RFP] plasmid. | ||
| - | We | + | The characterization was done in DH5alpha E.coli strain, which should not express TetR in normal conditions. We add increasing concentrations of ATC to these cells in order to evaluate to which extent these cells still express TetR; we also want to know the highest RFP expression we can get when Ptet is fully induced. |
| - | + | === ATC induction === | |
| - | The DH5alpha cells can still exhibit a basal level of TetR expression, even if the plasmid we transform doesn't contain TetR. By adding ATC, we are able to inhibit this basal expression of TetR, revealing the full power of Ptet as a promoter sequence. | + | ==== ''Description'' ==== |
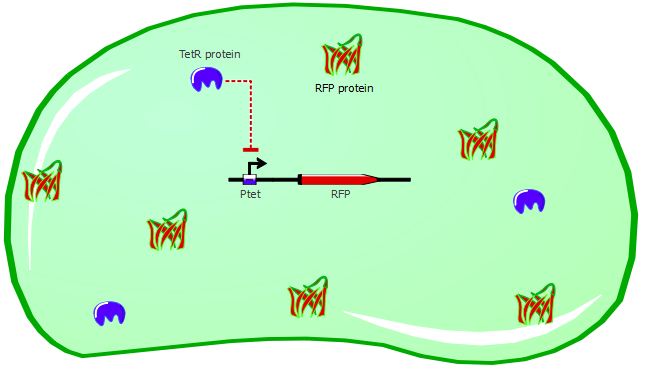
| + | The DH5alpha cells can still exhibit a basal level of TetR expression, even if the plasmid we transform doesn't contain TetR. By adding ATC, we are able to inhibit this basal expression of TetR, revealing the full power of Ptet as a promoter sequence. The action on ATC on our cells is depicted in the following cartoons: | ||
Cells without ATC: | Cells without ATC: | ||
| Line 20: | Line 21: | ||
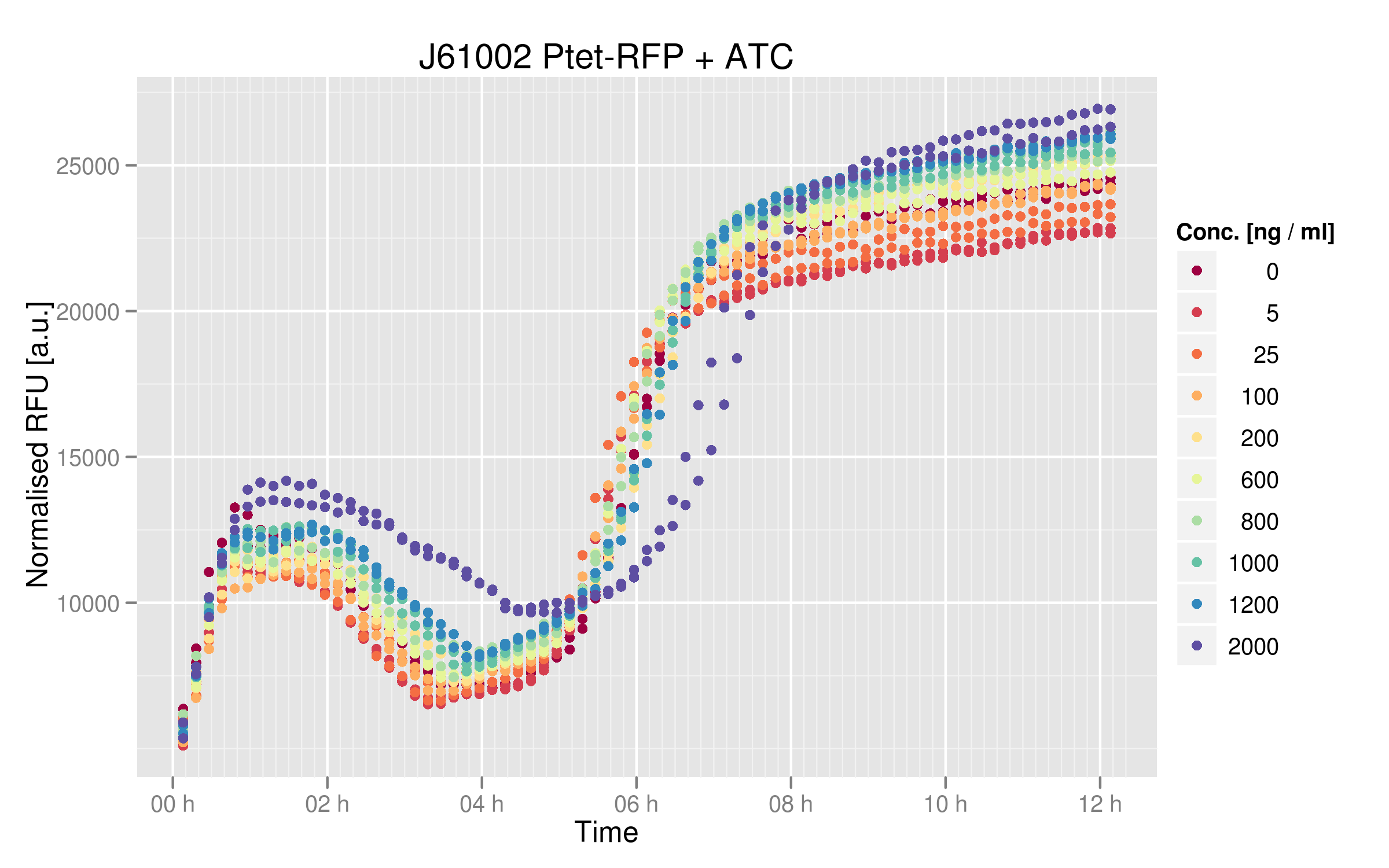
| - | Experimental results | + | ====''Experimental results''==== |
[[File:EPFL_Nadine-ptet-induction.png|600px]] | [[File:EPFL_Nadine-ptet-induction.png|600px]] | ||
| - | There is a clear difference between cells | + | There is a clear difference between cells not treated or treated with high doses of ATC. Still, this difference is not enormous, showing that our DH5alpha cells don't normally express much TetR. The basal expression of RFP is about 23'000 normalized RFUs, whereas with ATC we go up to 28'000 normalized RFUs. |
| - | + | ||
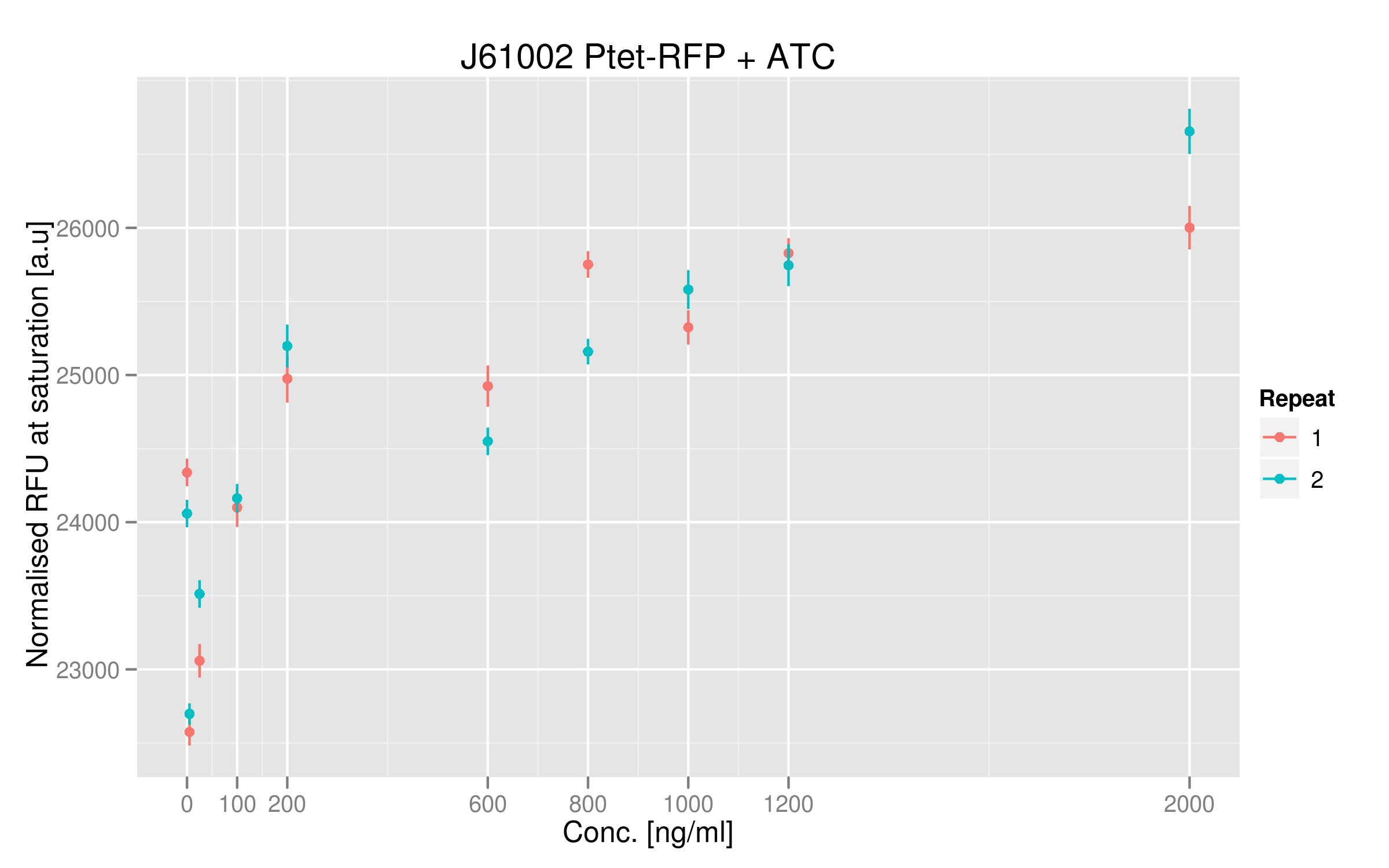
| + | === Dose-response curve === | ||
[[File:EPFL_Nadine-ptet-doseresponse.png|600px]] | [[File:EPFL_Nadine-ptet-doseresponse.png|600px]] | ||
Latest revision as of 20:11, 25 October 2011
In Vivo Characterization
In vivo Main | TetR-RFP System | TetR-LacI-RFP System | Ptet Characterization | Plac Characterization | Plasmid Details
Contents |
Ptet Characterization
Since we are using the Ptet promoter in combination with TetR in our reporter systems, it was useful for us to characterize the Ptet promoter only. This characterization was done with the J61002 Ptet-RFP plasmid.
The characterization was done in DH5alpha E.coli strain, which should not express TetR in normal conditions. We add increasing concentrations of ATC to these cells in order to evaluate to which extent these cells still express TetR; we also want to know the highest RFP expression we can get when Ptet is fully induced.
ATC induction
Description
The DH5alpha cells can still exhibit a basal level of TetR expression, even if the plasmid we transform doesn't contain TetR. By adding ATC, we are able to inhibit this basal expression of TetR, revealing the full power of Ptet as a promoter sequence. The action on ATC on our cells is depicted in the following cartoons:
Experimental results
There is a clear difference between cells not treated or treated with high doses of ATC. Still, this difference is not enormous, showing that our DH5alpha cells don't normally express much TetR. The basal expression of RFP is about 23'000 normalized RFUs, whereas with ATC we go up to 28'000 normalized RFUs.
Dose-response curve
The dose-response curve doesn't seem to saturate at the highest values of ATC added, indicating that some TetR proteins were perhaps still acting on the Ptet promoter. However, it is nice to see that TetR does have a quantifiable action on the promoter; it means that we can use it in our first readout system.
 "
"