Team:Arizona State/Project/L innocua
From 2011.igem.org
| Line 39: | Line 39: | ||
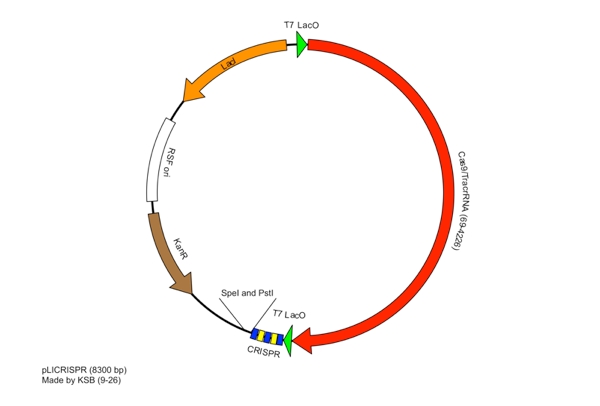
<p>The Plasmid conferring the LIsteria Innocua based CRISPR was assembled in one ligation step. Cas9 (with the tracrRNA region) amplified from the Listeria innocua CLIP11262 (Link to ATCC) genome using primers that incorporated NcoI and NotI sites. The array, including a T7 Promoter to drive transcription and the leader sequence directly adjacent to the CRISPR repeats and spacers, was amplified from its pIDTSmart plasmid using M13 Forward (-20) and M13 Reverse (-27). The PCR products where then digested with NcoI and NotI (for Cas9), NotI and XhoI for the CRISPR array. The vector, pRSFDuet-1 (Novagen), was cut with NcoI and XhoI and dephosphorylated. Both inserts and vector were combined with the vector, ligated and transformed. This method allows for the formation of a functional CRISPR array in one cloning step.</p> | <p>The Plasmid conferring the LIsteria Innocua based CRISPR was assembled in one ligation step. Cas9 (with the tracrRNA region) amplified from the Listeria innocua CLIP11262 (Link to ATCC) genome using primers that incorporated NcoI and NotI sites. The array, including a T7 Promoter to drive transcription and the leader sequence directly adjacent to the CRISPR repeats and spacers, was amplified from its pIDTSmart plasmid using M13 Forward (-20) and M13 Reverse (-27). The PCR products where then digested with NcoI and NotI (for Cas9), NotI and XhoI for the CRISPR array. The vector, pRSFDuet-1 (Novagen), was cut with NcoI and XhoI and dephosphorylated. Both inserts and vector were combined with the vector, ligated and transformed. This method allows for the formation of a functional CRISPR array in one cloning step.</p> | ||
| - | [[Image: ASU_Listeria_Gel_9-28.PNG| | + | [[Image: ASU_Listeria_Gel_9-28.PNG|800px]] |
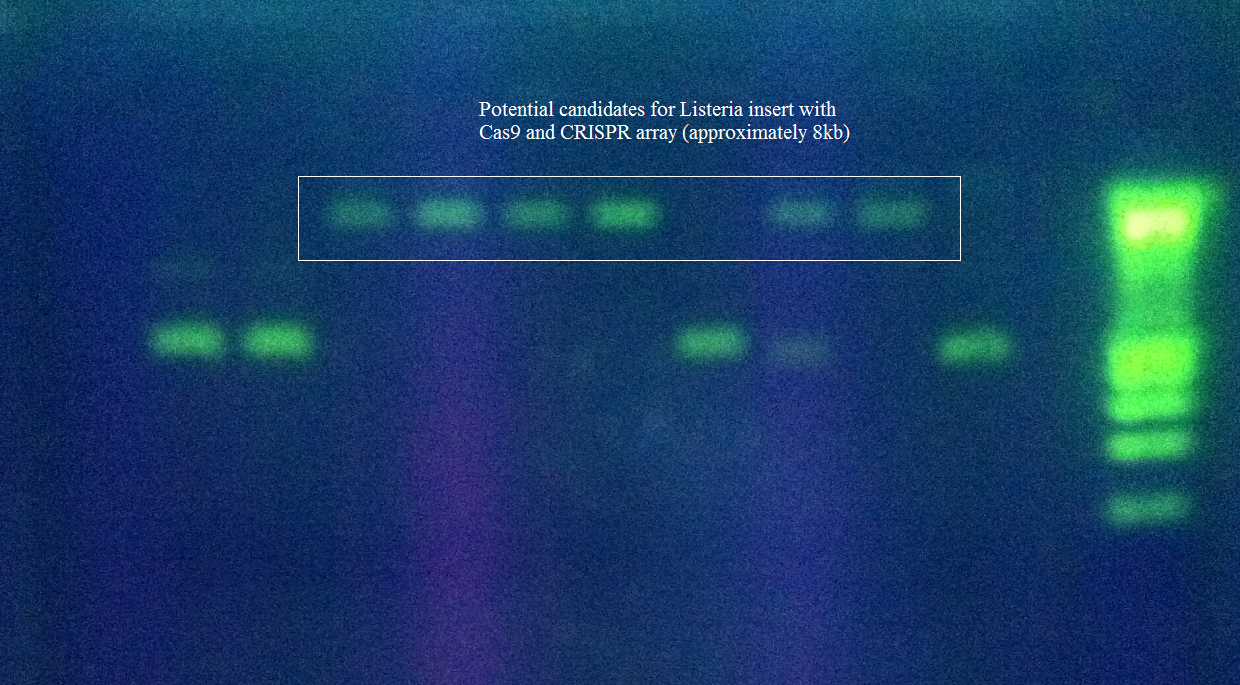
<p>Transformants were screened for the correct size of insert by PCR from the plasmid and linearization of the plasmid and visualization of size on 1% agarose gel.</p> | <p>Transformants were screened for the correct size of insert by PCR from the plasmid and linearization of the plasmid and visualization of size on 1% agarose gel.</p> | ||
Revision as of 03:38, 29 September 2011
|
|
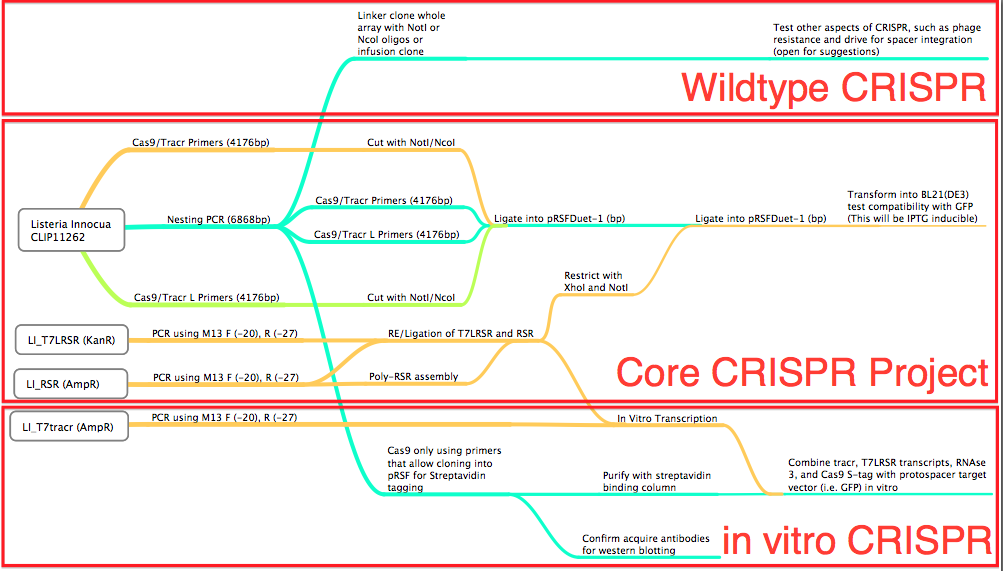
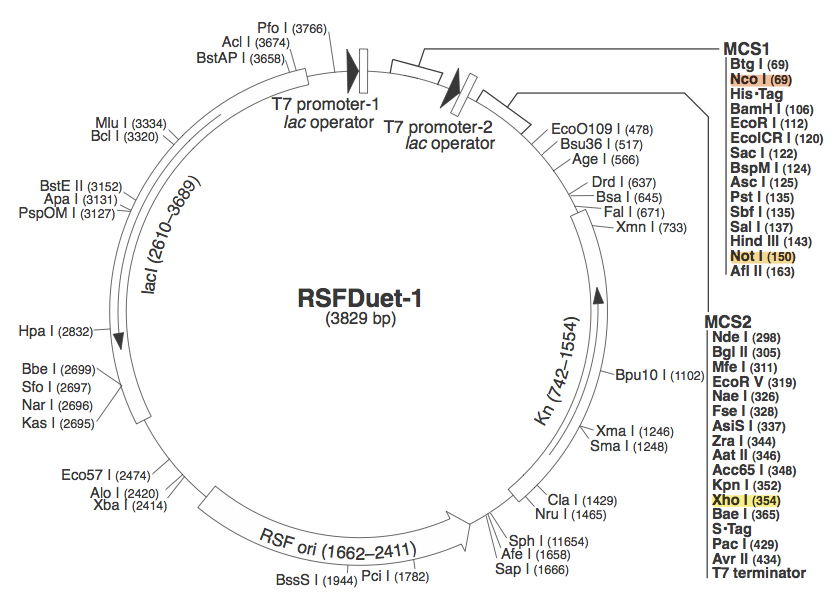
Conception: Why Listeria Innocua?BioBrickable ImmunityThrough the development of the multiple CRISPR projects for the our iGEM team, we realized the need to reduce the number of cloning steps, decrease the size of the CRISPR associated genes needed, and to allow for a robust method to construct the CRISPR array. Listeria Innocua, an organism with a Type 2 CRISPR[62][65], utilizes only Cas9 and a small Trans-encoded CRISPR RNA (tracrRNA), with endogenous RNase3 to prepare mature CRISPR RNA and exert CRISPR mediated endonuclease activity. This organism in effect allows for a much more compact CRISPR plasmid to be constructed compared to the E.coli (Type 1) CRISPR pathway that amounts to 8Kb of coding sequences for functionality. Listeria overcomes this conundrum. Reconstructing the Listeria innocua CRISPR pathway on a plasmid requires only 4Kb and is compatible with the BioBrick Suffix (SpeI and PstI) sites. Better Regulation of the PathwayOrganisms, like BL21(DE3), that are ideal for protein expression do not produce Ion and OmpT proteases, which are responsible for intracellular and extracellular protein degradation. The lack of these two protease genes allows for the formation of stable recombinant proteins, but also means the proteins have a long half-life, thus once the CAS proteins production is induced, they remain active for a long time. For BL21(DE3) and other like organisms to be the host of a tightly controllable CRISPR pathway a non-protein, RNA based, mechanism for regulation of Mature CRISPR RNA (crRNA) formation is required. The CRISPR pathway in Listeria Innocua, and other organisms of the same CRISPR subtype, utilize a trans-encoded CRISPR RNA (tracrRNA) to initiate the formation of mature CRISPR RNA (crRNA) (62). The tracrRNA allows for a greater regulation of the mature CRISPR RNA formation, reducing the amount of leaky expression and allowing for faster toggling of CRISPR active and inactive states. More immunity optionsCRISPR based immunity requires complete sequence complimentarily between the CRISPR generated spacer and proto-spacer (Target) seed sequence and Proto-spacer Adjacent Motif (PAM)[70][74]. The PAM sequence required for CRISPR function and is conserved for different organisms. For instance: E.coli’s PAM sequence is ATG, meaning an ATG must be directly adjacent to the 5’ end of the proto-spacer[70]. Streptococcus Thermophilus’ PAM sequence is NNAGAAW or NGGNG at the 3’ end of the target, proto-spacer[74]. Listeria Innocua’s PAM is only NGG[30], which means a large target sequence possesses 4 times the number of potential proto-spacers compared to E.coli and approximately 16 times compared to Streptococcus thermophilus. DesignFlowchartThis is our experimental flowchart for the L. innocua CRISPR platform. pRSF DuetWe chose to use pRSF Duet from Novagen as the vector for our construct because it has two multiple cloning sites (MCS1 and MCS2). It was used in a CRISPR study (cite, is this Brouns?). In the image above, the restriction sites for each component of our construct are highlighted. Red --> Orange - Cas 9
These restriction sites were specifically chosen because they do not fall within the Cas, Leader, or RSR sequences. This was confirmed using [http://tools.neb.com/NEBcutter2/ NEB Cutter]. Our PlasmidThe plasmid containing CRISPR derived from Listeria Innocua CLIP11262 contains the standard BioBrick Suffix (SpeI and PstI Sites) at the end of the array distal to the Leader sequence and T7 Promoter, thus Biobrick compatible Repeat Spacer Repeat (RSR) sections can sequentially be added into the vector to expand the CRISPR repertoire. ConstructionThe Plasmid conferring the LIsteria Innocua based CRISPR was assembled in one ligation step. Cas9 (with the tracrRNA region) amplified from the Listeria innocua CLIP11262 (Link to ATCC) genome using primers that incorporated NcoI and NotI sites. The array, including a T7 Promoter to drive transcription and the leader sequence directly adjacent to the CRISPR repeats and spacers, was amplified from its pIDTSmart plasmid using M13 Forward (-20) and M13 Reverse (-27). The PCR products where then digested with NcoI and NotI (for Cas9), NotI and XhoI for the CRISPR array. The vector, pRSFDuet-1 (Novagen), was cut with NcoI and XhoI and dephosphorylated. Both inserts and vector were combined with the vector, ligated and transformed. This method allows for the formation of a functional CRISPR array in one cloning step. Transformants were screened for the correct size of insert by PCR from the plasmid and linearization of the plasmid and visualization of size on 1% agarose gel. Future Applications of Listeria-based CRISPRCRISPR/Cas pathways based on E.coli requires a large compliment of cas genes (ie. CasABCDE and Cas3). This means multiple cloning steps, and it does not lend itself to unlimited expansion of the CRISPR array. Listeria innocua potentially forgoes the downfalls of other CRISPR pathways. Cas9 is the only CRISPR associated structural gene required for immunity. The trans-encoded CRISPR RNA (tracrRNA) potentially allows for greater regulation and rapid toggling of CRISPR function. The interaction of the tracrRNA and the CRISPR RNA is required for the formation of functional CRISPR RNA (62), thus, if tracrRNA is not transcribed, then mature RNA cannot be formed, abrogating CRISPR activity. The Listeria CRISPR construct is compatible with insertion of BioBricks on the distal end of the CRISPR array, thus any Repeat Spacer Repeat (RSR) formatted BioBrick cut with XbaI and PstI can be inserted into the plasmid backbone of listeria’s CRISPR that is cut with SpeI and PstI. Listeria’s small proto-spacer adjacent motif (PAM) sequence increases the likelihood of finding potential targets over those of E.coli and Streptococcus Thermophilus several-fold. The combination of these advantages make the CRISPR/Cas pathway of Listeria Innocua not just another ‘CRISPR ohlala’ project. This project attempts to make CRISPR-based immunity and the ability to encompass a diverse repertoire of spacers accessible to every synthetic biologist. |
 "
"