Team:Arizona State/Project/B halodurans
From 2011.igem.org
| Line 3: | Line 3: | ||
==Conception== | ==Conception== | ||
| - | ( | + | Besides the DNA-targeting Crispr-Cas system in E. coli, our other original project was the construction of an RNA-targeting Crispr-Cas system from Bacillus halodurans. Early attempts to explain the function of the Crispr-Cas system proposed that the Crispr array was utilized to target RNA, not DNA, in a manner analogous to eukaryotic RNAi (Makarova et al, 2006). While later studies showed that DNA, not RNA, was the target of certain Crispr-Cas systems, at least one study found a Crispr-Cas system that seemed to target RNA (Marrafini and Sontheimer, 2008; Hale et al, 2009). This RNA-targeting CRISPR-Cas system used so-called repeat associated mysterious proteins (RAMPs) in the form of CMRs 1-6. |
| + | We realized the potential importance of the successful creation and characterization of such an RNA targeting Crispr-Cas system and thought it would complement our E. coli-based DNA targeting system well. We chose B. halodurans because of its availability and because its CMR1-6 proteins are localized together on its genome, similar to the successful RNA-targeting CRISPR-Cas system (Haft et al, 2005). | ||
| + | |||
==Design== | ==Design== | ||
Revision as of 23:53, 28 September 2011
|
|
ConceptionBesides the DNA-targeting Crispr-Cas system in E. coli, our other original project was the construction of an RNA-targeting Crispr-Cas system from Bacillus halodurans. Early attempts to explain the function of the Crispr-Cas system proposed that the Crispr array was utilized to target RNA, not DNA, in a manner analogous to eukaryotic RNAi (Makarova et al, 2006). While later studies showed that DNA, not RNA, was the target of certain Crispr-Cas systems, at least one study found a Crispr-Cas system that seemed to target RNA (Marrafini and Sontheimer, 2008; Hale et al, 2009). This RNA-targeting CRISPR-Cas system used so-called repeat associated mysterious proteins (RAMPs) in the form of CMRs 1-6. We realized the potential importance of the successful creation and characterization of such an RNA targeting Crispr-Cas system and thought it would complement our E. coli-based DNA targeting system well. We chose B. halodurans because of its availability and because its CMR1-6 proteins are localized together on its genome, similar to the successful RNA-targeting CRISPR-Cas system (Haft et al, 2005).
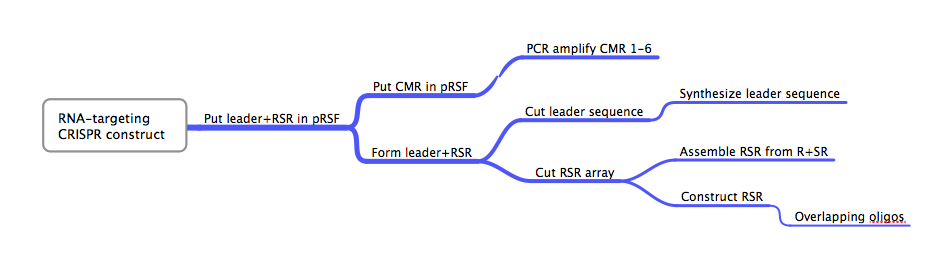
DesignFlowchartThis is the experimental flowchart for the construction of our B. halodurans CRISPR platform. Beginning from the right-hand side of the diagram, this image outlines each step of the construction of our synthetically directed, RNA-targeting B. halodurans CRISPR-Cas construct. In short: Step 1) Construct RSR arrays with one of two methods:
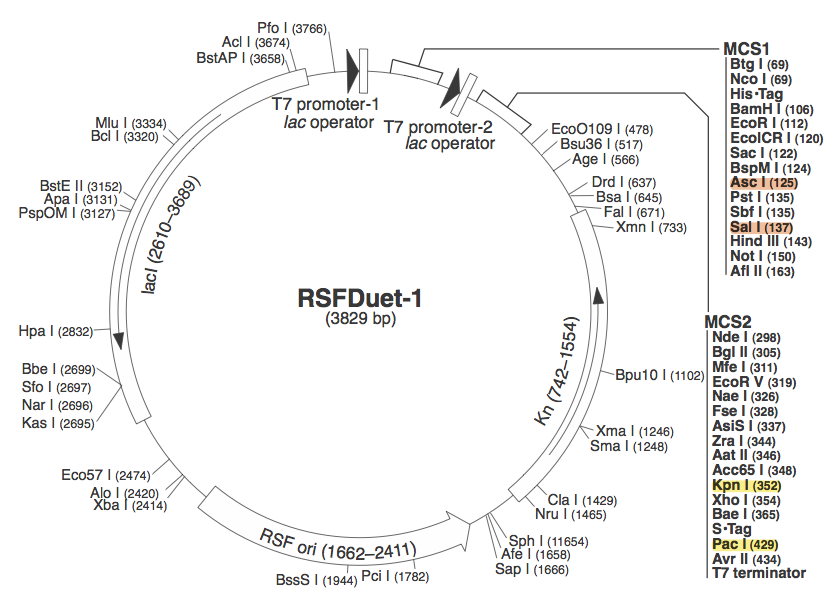
pRSF DuetWe chose to use pRSF Duet from Novagen as the vector for our construct because it has two multiple cloning sites (MCS1 and MCS2). It was used in a CRISPR study (cite, is this Brouns?). In the image above, the restriction sites for each component of our construct are highlighted.
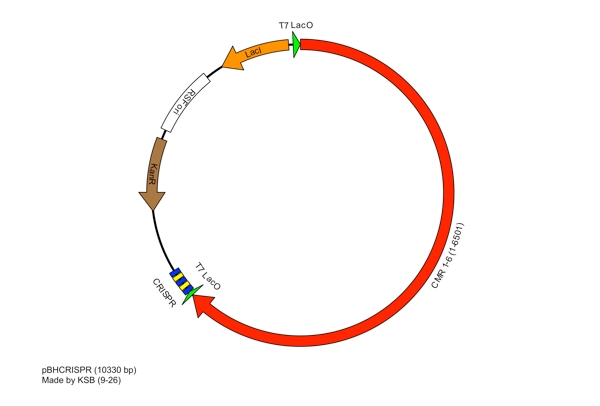
These restriction sites were specifically chosen because they do not fall within the Cas, Leader, or RSR sequences. This was confirmed using [http://tools.neb.com/NEBcutter2/ NEB Cutter]. Our PlasmidCompleting each of these steps yields this plasmid: The plasmid containing CRISPR derived from Bacillus halodurans includes the CMR region. In addition, it includes a leader sequence corresponding to B. halodurans CRISPR constructs, which is adjacent to our GFP-targeting Repeat/Spacer array. ConstructionRepeat-Spacer ArrayOriginally, we envisioned the Repeat-Spacer-Repeat (RSR) array as an easily manipulatable, modular, BioBrick-compatible component with restriction sites flanking the spacer. This would mean that once a functional CRISPR construct is constructed, all it would take is a simple restriction and ligation to insert a spacer of one's choice that corresponds to a gene of one's choice. Thus, we constructed two potential BioBricks: "RA": BB Prefix, Restriction Site #1 inside anti-GFP Spacer, Restriction Site #2, B. halodurans Repeat Sequence, Suffix
The idea here was to connect multiple RA inserts and cap the construct with a RB insert, effectively creating an array of any given length. However, upon further examination of CRISPR literature, it was hypothesized that the scars left by restrictions and ligations would impede proper hairpinning and Cascade interaction of transcribed cRNA. So, we then took a different and more budget-friendly assembly approach: overlapping oligos. (more here) Leader+RSRDiscuss leader (BioBrick), any assembly? Cas genesSuccess! (image+caption) FutureAdvantages of RNA-targeting CRISPRLots of potential cool applications of this... list them? Overcoming Tandem Repeat IssuesDiscuss this, similar to E. coli... do overlapping oligos solve this at all? |
 "
"