Team:Washington/Magnetosomes/Magnet Results
From 2011.igem.org
(→mamK: Filament formation) |
(→Our favorite genes in pGA vectors: Magnetosome gene-protein Fusions) |
||
| Line 17: | Line 17: | ||
(for other gene functions, please see the iGEM Toolkits parts submitted page)Using our two genes of interest, we created C-terminal sfGFP fusions so we could track the localization of each gene separately within ''E.coli.'' | (for other gene functions, please see the iGEM Toolkits parts submitted page)Using our two genes of interest, we created C-terminal sfGFP fusions so we could track the localization of each gene separately within ''E.coli.'' | ||
| - | + | ===mamK: Filament formation=== | |
| + | <br> | ||
<center> [[File:Washington igem11 MamK fusion full 01.jpg|400px|middle]][[File:Washington igem11 MamK fusion gfp 01.jpg||400px|middle]]</center> | <center> [[File:Washington igem11 MamK fusion full 01.jpg|400px|middle]][[File:Washington igem11 MamK fusion gfp 01.jpg||400px|middle]]</center> | ||
| Line 33: | Line 34: | ||
</gallery> | </gallery> | ||
| - | <br> | + | <br> |
| - | + | ||
===mamI: Membrane Localization=== | ===mamI: Membrane Localization=== | ||
Revision as of 22:46, 28 September 2011
What’s in the Magnetosome Toolkit?
- Our favorite genes in pGA vectors
- A set of the 18 essential genes for the various steps of magnetosome formation
- A table compiling individual gene functions from our literature search
Our favorite genes in pGA vectors: Magnetosome gene-protein Fusions
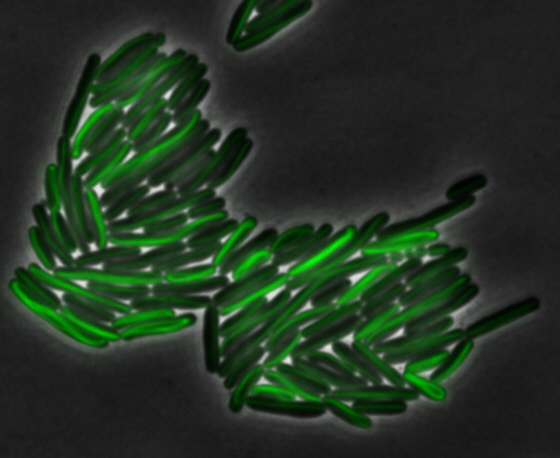
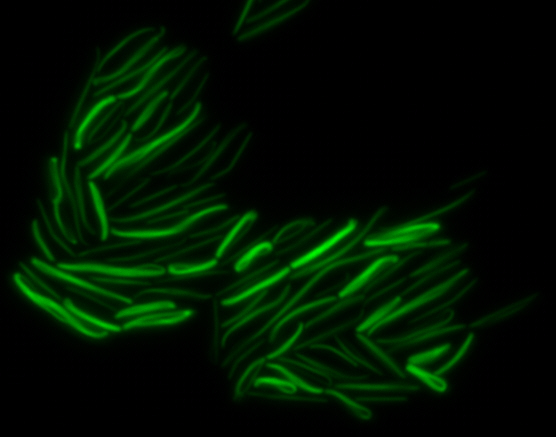
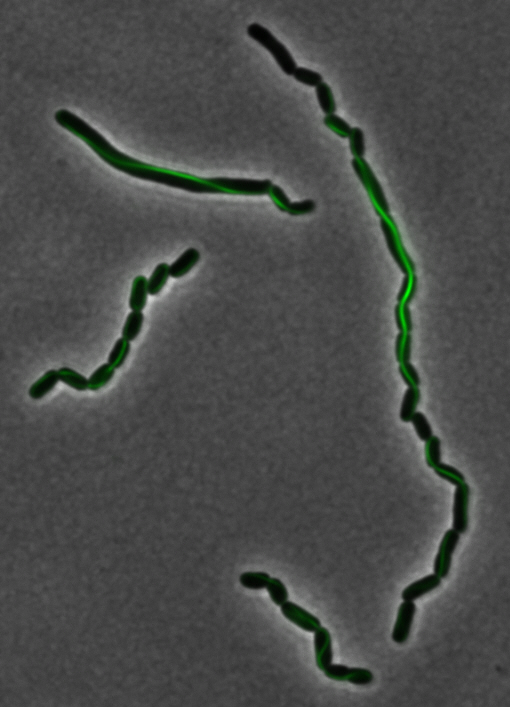
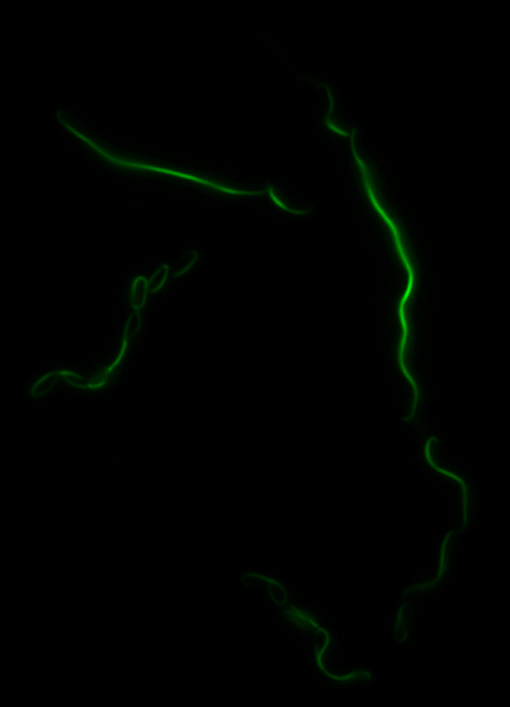
As previously stated, our genes of interest were mamK and mamI as they have functions related to localization of the magnetosome. Specifically, mamK is a bacterial actin-like cytoskeleton protein required for proper alignment of the magnetosomes in a chain. mamK is also shown to localize the mamI, which loss of mamK inhibits membrane formation. (for other gene functions, please see the iGEM Toolkits parts submitted page)Using our two genes of interest, we created C-terminal sfGFP fusions so we could track the localization of each gene separately within E.coli.
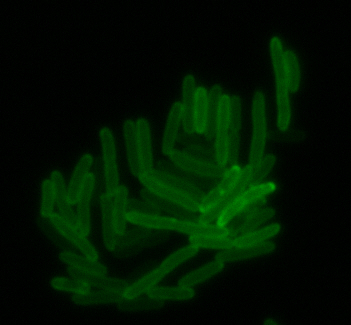
mamK: Filament formation
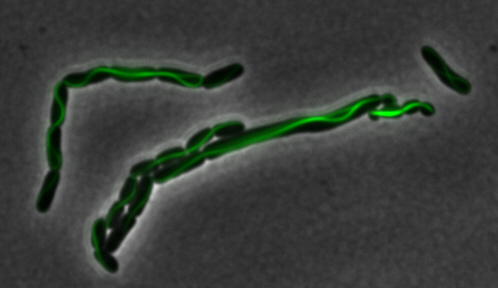
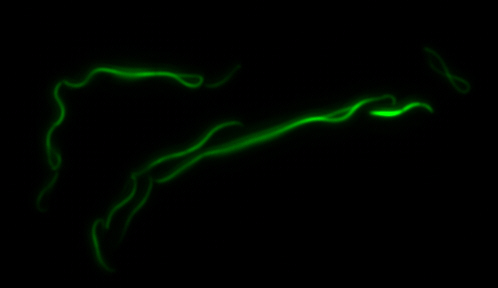
The results we obtained with our sfGFP fusions inside E.coli were comparable to those done through other studies in the host organism Magnetospirillum magneticum. Within AMB-1, mamK is a filament which runs through the length of the bacteria. In the our images of mamK, filamentous structures can be clearly seen running through the length of many bacteria. In our experimental result, there was an over- expression of mamK which connected the E.coli cells together.
mamI: Membrane Localization
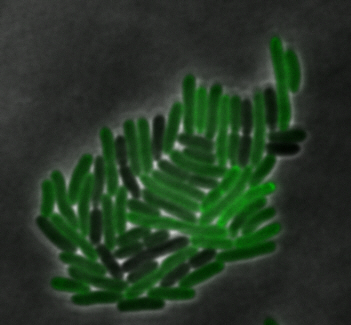
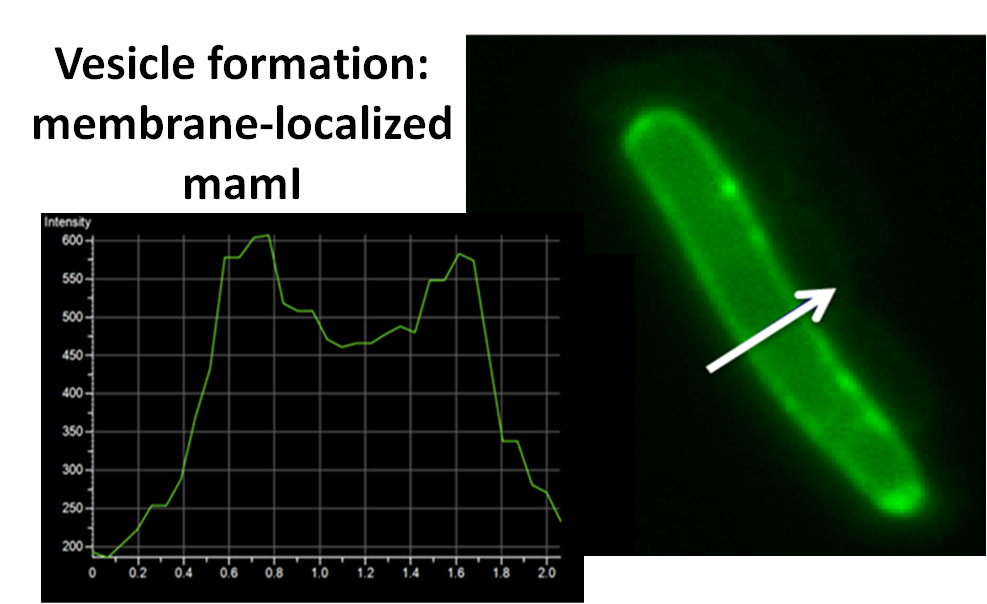
For mamI, the gene product is seen to fluorescent around the bacterial cell membrane of the bacteria but mostly concentrated at the ends. This can be seen in the fluorescence profile analysis that was taken while imaging the cells. The graph shows that as the arrow crosses the cell membrane, the fluorescent peaks are at a maximum, and through the center of the cell, the level of fluorescence decreases.
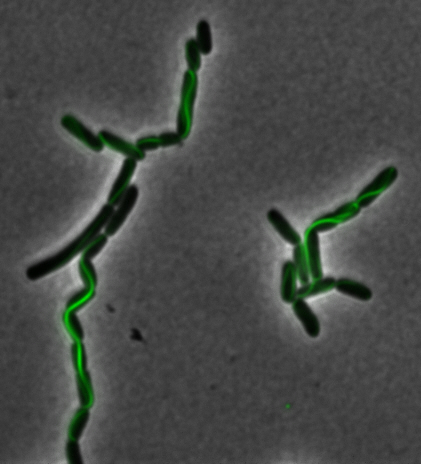
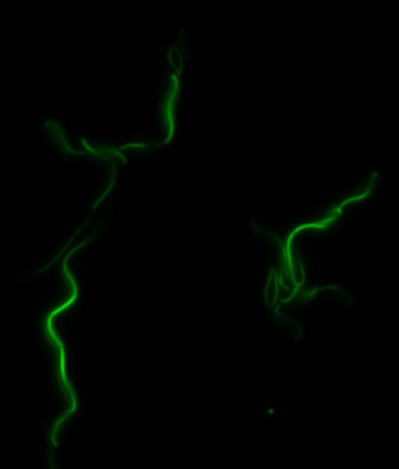
Construction of the R5 region of the Magnetosome Island in E.coli
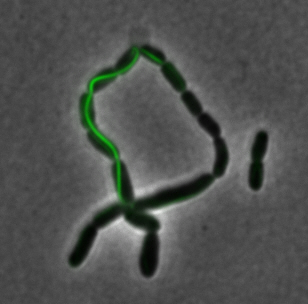
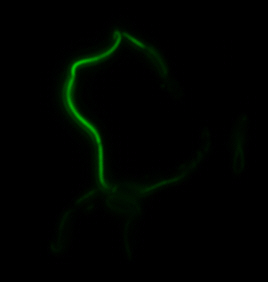
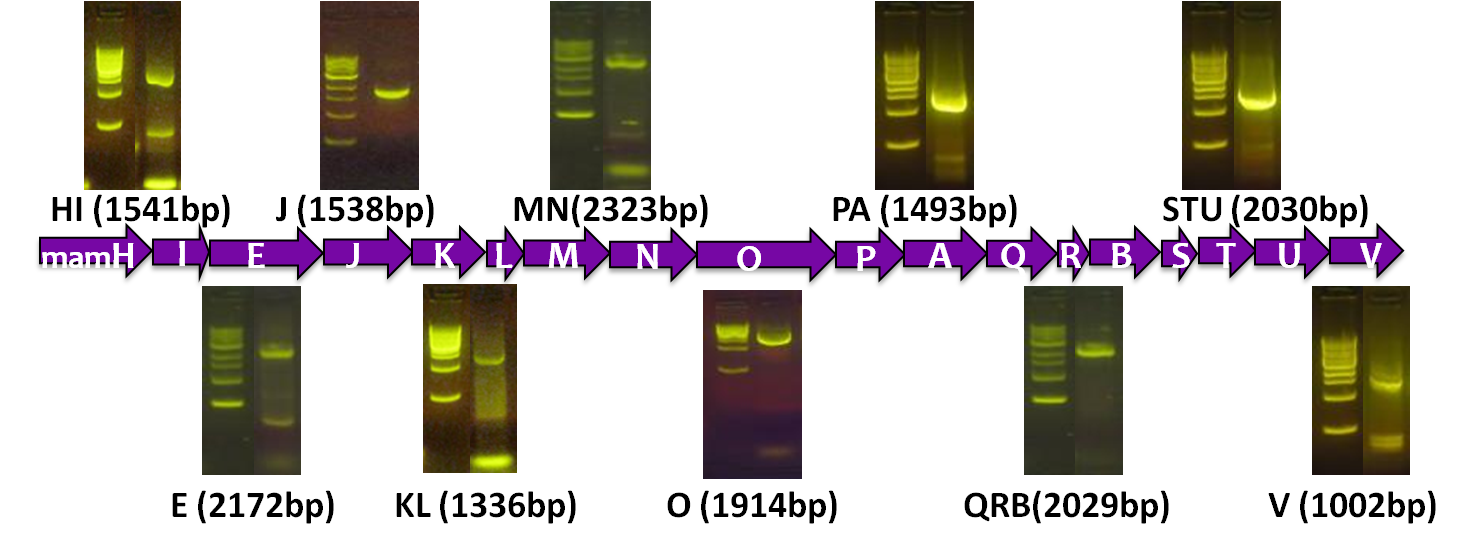
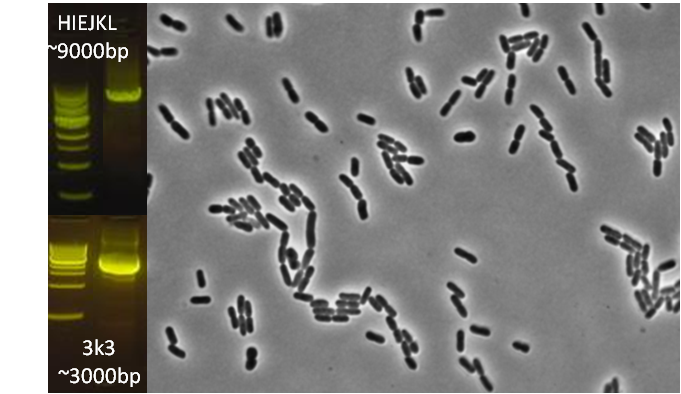
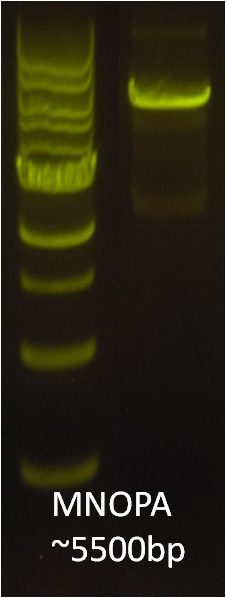
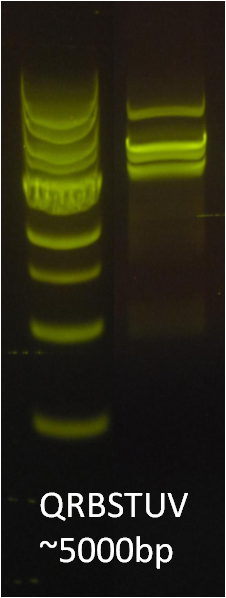
After identifying that the construction of the scaffold had worked, we proceeded to work on the final assembly in three parts: mamHIEJKK, mamMNOPA, and mamQRBSTUV. The PCR products of the first, and the third part of the assembly are shown below. Both fragments of the assembly have been partially sequenced confirmed, and we are currently working on designing primers to fill in the gap sequences. Despite these gaps, when this samples were imaged, filaments in the first part (mamHIEJKL)were still apparent.
 :
:
A set of the 18 essential genes for the various steps of magnetosome formation
Before piecing together the 16 kb genome of the mamAB gene cluster within the magnetosome island (MAI), we extracted out the genes in the following groups:
| Gene groups | Length (bp) |
|---|---|
| mamHI | 1541 |
| mamE | 2172 |
| mamJ | 1538 |
| mamKL | 1336 |
| mamMN | 2323 |
| mamO | 1914 |
| mamPA | 1493 |
| mamQRB | 2029 |
| mamSTU | 2030 |
| mamV | 1002 |
A table of individual gene functions
Please see the bottom of our parts submitted page.
 "
"