Team:Arizona State/Project/E coli
From 2011.igem.org
| Line 1: | Line 1: | ||
{{:Team:Arizona State/Templates/main|title=E. coli|content= | {{:Team:Arizona State/Templates/main|title=E. coli|content= | ||
__NOTOC__ | __NOTOC__ | ||
| + | |||
| + | ==Conception== | ||
| + | |||
| + | <p>Our first foray into manipulating CRISPR was in E. coli. We chose to use the CRISPR-Cas system of K12-MG1655 because the strain is well-characterized and it is a common lab organism, making it easy to obtain. Its ubiquity would allow for a manipulatable CRISPR platform useful in many lab applications.</p> | ||
| + | |||
| + | <p>Another reason for seeking to synthetically direct CRISPR in E. Coli is the existence of strains that do not have Cas genes. For instance, BL21 DE3 does not have a natural CRISPR-Cas system, making it an ideal organism for the transfer of our CRISPR construct. In short, we would obtain Cas genes from MG1655, insert them into a plasmid with a Repeat-Spacer array that corresponds to GFP, and then transfer that plasmid into the BL21 DE3 host along with a constitutive GFP plasmid. If successful, the expression of GFP would be silenced or significantly decreased. </p> | ||
| + | |||
| + | <p>Because there are E. coli strains with Cas genes and strains without Cas genes, one can easily remove Cas genes that will integrate new spacers. In this case, Cas 1 and Cas 2 were excluded, as they have been shown in literature to facilitate new spacer integration (cite). In order to properly test our CRISPR construct and ensure that silencing of GFP is a result of the addition of our pCRISPR, it is important to prevent other spacers from being added to the Repeat-Spacer array.</p> | ||
==Experimental Design== | ==Experimental Design== | ||
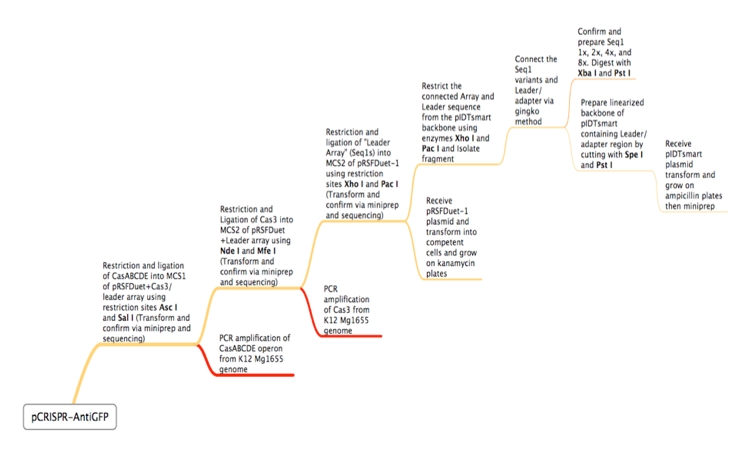
| - | This is | + | This is the experimental flowchart for the construction of our E. coli CRISPR platform. |
[[Image: ASU_1655_flowchart.jpg]] | [[Image: ASU_1655_flowchart.jpg]] | ||
| - | + | Beginning from the right-hand side of the diagram, this image outlines each step of the construction of our synthetically directed, GFP-targeting E. coli CRISPR-Cas construct. In short: | |
| + | |||
| + | <br> Step 1) Construct RSR arrays with increasing numbers of anti-GFP spacers (1x, 2x, 4x, 8x) | ||
| + | <br> Step 2) PCR amplify Cas genes (CasABCDE, Cas3) | ||
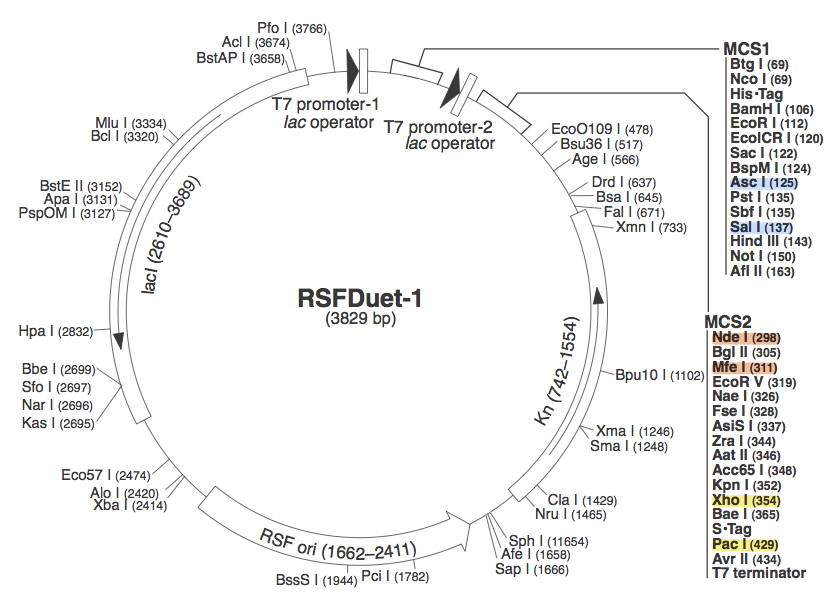
| + | <br> Step 3) Insert both the array and the Cas genes into plasmid with two multiple cloning sites (pRSF Duet) | ||
| + | |||
| + | [[Image: ASU_pRSF.png]] | ||
| + | |||
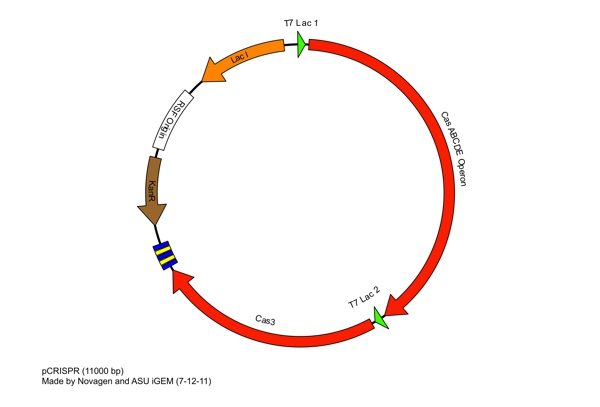
| + | Completing each of these steps yields this plasmid: | ||
[[Image: ASU_pK12CRISPR.jpg]] | [[Image: ASU_pK12CRISPR.jpg]] | ||
| - | The plasmid | + | The plasmid design for the Anti-GFP CRISPR construct in E. coli includes Cas genes A, B, C, D, E, and 3. In addition, it includes a leader sequence corresponding to most K12 CRISPR constructs, which is adjacent to our GFP-targeting Repeat/Spacer array and promotes its expression. |
| + | |||
| + | ==Construction== | ||
| + | |||
| + | <b>Repeat-Spacer-Repeat Array</b> | ||
| + | |||
}} <!-- close content attribute for menubar --> | }} <!-- close content attribute for menubar --> | ||
Revision as of 11:26, 28 September 2011
|
|
ConceptionOur first foray into manipulating CRISPR was in E. coli. We chose to use the CRISPR-Cas system of K12-MG1655 because the strain is well-characterized and it is a common lab organism, making it easy to obtain. Its ubiquity would allow for a manipulatable CRISPR platform useful in many lab applications. Another reason for seeking to synthetically direct CRISPR in E. Coli is the existence of strains that do not have Cas genes. For instance, BL21 DE3 does not have a natural CRISPR-Cas system, making it an ideal organism for the transfer of our CRISPR construct. In short, we would obtain Cas genes from MG1655, insert them into a plasmid with a Repeat-Spacer array that corresponds to GFP, and then transfer that plasmid into the BL21 DE3 host along with a constitutive GFP plasmid. If successful, the expression of GFP would be silenced or significantly decreased. Because there are E. coli strains with Cas genes and strains without Cas genes, one can easily remove Cas genes that will integrate new spacers. In this case, Cas 1 and Cas 2 were excluded, as they have been shown in literature to facilitate new spacer integration (cite). In order to properly test our CRISPR construct and ensure that silencing of GFP is a result of the addition of our pCRISPR, it is important to prevent other spacers from being added to the Repeat-Spacer array. Experimental DesignThis is the experimental flowchart for the construction of our E. coli CRISPR platform. Beginning from the right-hand side of the diagram, this image outlines each step of the construction of our synthetically directed, GFP-targeting E. coli CRISPR-Cas construct. In short:
Completing each of these steps yields this plasmid: The plasmid design for the Anti-GFP CRISPR construct in E. coli includes Cas genes A, B, C, D, E, and 3. In addition, it includes a leader sequence corresponding to most K12 CRISPR constructs, which is adjacent to our GFP-targeting Repeat/Spacer array and promotes its expression. ConstructionRepeat-Spacer-Repeat Array |
 "
"