From 2011.igem.org
(Difference between revisions)
|
|
| Line 1: |
Line 1: |
| | {{:Team:Harvard/Template:PracticeBar2}} | | {{:Team:Harvard/Template:PracticeBar2}} |
| - | <div class="whitebox2">
| + | {{:Team:Harvard/Template:ResultsGrayBar}} |
| - | [[Team:Harvard/Results | Overview]] | [[Team:Harvard/Results/MAGE | MAGE]] | [[Team:Harvard/Results/Lambda_Red | Lambda Red]]| [[Team:Harvard/Results/Chip_Synthesis | Chip-Based Library]] | [[Team:Harvard/Results/One-Hybrid_Selection | One-Hybrid Selection]] | [[Team:Harvard/Results/Zinc_Finger_Binders | Zinc Finger Binders]] | [[Team:Harvard/Results/Biobricks | Biobricks]]
| + | |
| - | </div>
| + | |
| | <div class="whitebox"> | | <div class="whitebox"> |
| | __NOTOC__ | | __NOTOC__ |
Revision as of 00:43, 26 September 2011
bar
qPCR curves
When performing a qPCR, one wants to run the reaction while the growth is still exponential. During this phase, all the oligos are replicated at equal rates. Once growth levels off, the reaction should be stopped as the oligos are now being replicated unequally. Each of the graphs below represents a qPCR for one of our sub-pools.
| CB Top |
| CB Bot |
| FH Top |
| FH Bot |
| Myc 198 |
| Myc 981 |
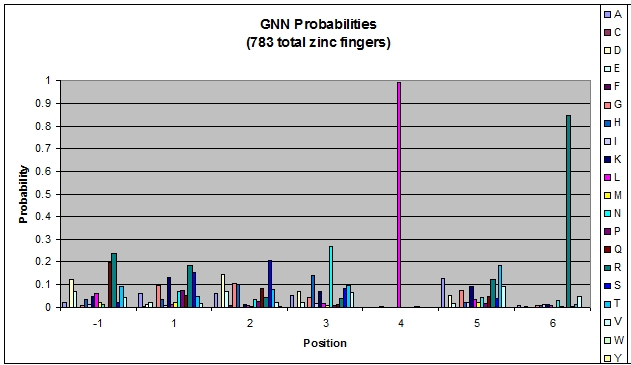
Probability data for the 783 fingers that bind to GNN triplets. Note the high probability of leucine at position 4 and arginine at position 6.
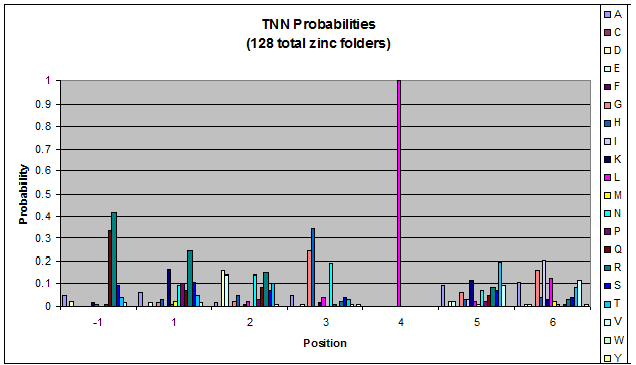
Probability data for the 128 fingers that bind to TNN triplets. Note the high probability of leucine at position 4.
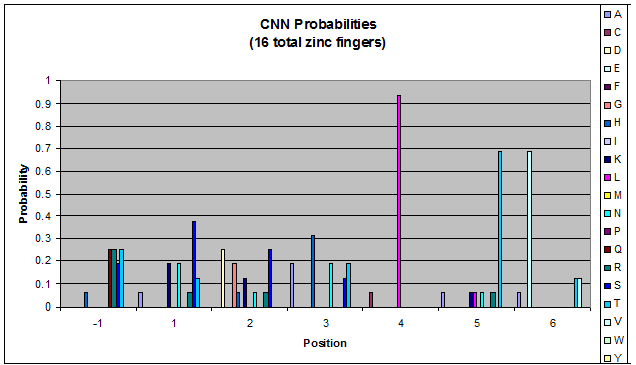
Probability data for the 16 fingers that bind to CNN triplets. There may not be enough data to consider this information statistically significant
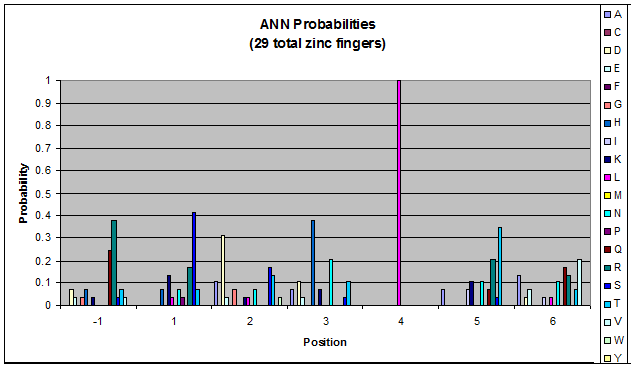
Probability data for the 29 fingers that bind to ANN triplets. There may not be enough data to consider this information statistically significant
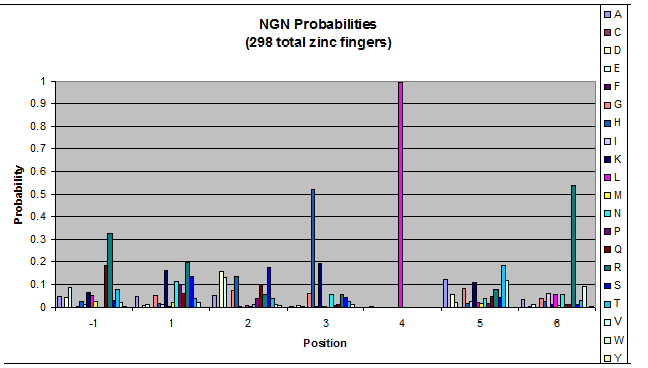
Probability data for the 298 fingers that bind to NGN triplets. The position 4 leucine motif remains. There is also a high probability (> 0.5) of a histidine at position 3 and an arginine at position 6.
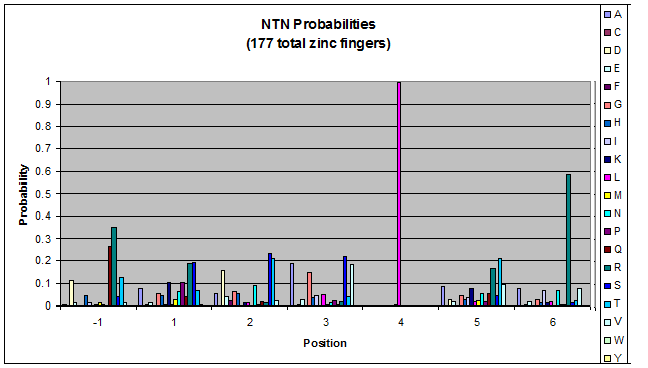
Probability data for the 177 fingers that bind to NTN triplets. The position 4 leucine motif remains.
Sequencing results of Library transformation
Error rates
Still being updated
- Good: 57.8% (22/38)
- Single SNP: 5.3% (2/38)
- Multiple SNPs: 18.4% (7/38)
- Frame shift: 18.4% (7/38)
Distributions
Still being updated
Each of the sequenced zinc fingers have original F1 fingers.
 "
"














