green light receptor
PCR to amplify Green light Promotor PcpcG
Investigators:Julia
PCR
| Name: Julia
| Date: 29.08.2011
|
| Project Name: Amplification of Green light Promotor
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
|
|
| 10µl
| 5x Phusion Buffer
|
|
| 2.5µl
| Primer fw
| tatgaattcgcggccgcttctagaCCATTGTGCTTTTCTCTATCAACC
|
| 2.5µl
| Primer dw
| tatctgcagcggccgctactagtaACTTAAAAGTTGTTTAATGTCCAGCC
|
| 1µl
| dNTPs
|
|
| 1µl
| DNA-Template
| original Synechocystis genome
|
| 0.5 µl
| Phusion
|
|
What program do you use?
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
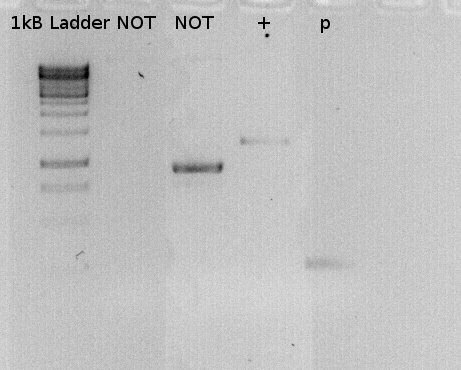
the lane labeled P, is the PCR product.Expected size: 264 bp
Testdigest of quickchanged CcaR and Precipitator
Investigators:Julia,with help of the team
3μl of the Miniprep were digested with EcoRI and PstI.

we are going to sequence the Precipitator samples 61a and b
(wrong labeling on the pic, its the 63 in the right corner)
62 b and f
63 c and d
and of the green light response regulator qCcaR 17b and 18a
blue light receptor
3A-assembly
Investigators: Sandra
Digestion I:
- Lovtap
- Not-Gate
- pSB1A3
- pSB1T3
stored in the blue light box.
Ligation
- ♥-A3 Not
- ♥-A3 nOt
- ♥-A3 noT
- ♥-T3 Not
- ♥-T3 nOt
- ♥-T3 noT
stored in the ligation box.
Digestion II:
Also digestion with new amplified vector
Lovtap was digested again this time also with DpnI. So for this ligation the ♥-PCR3A, digested with DpnI, and pSB1C3 (new amplified with PCR, already digested with DpnI) and Not, nOt, noT were used.
- ♥-C3 Not
- ♥-C3 nOt
- ♥-C3 noT
Transformation
Investigators: Sophie
transformation of:
- ♥-A3 Not
- ♥-A3 nOt
- ♥-A3 noT
- ♥-T3 Not
- ♥-T3 nOt
- ♥-T3 noT
Incubation over night at 37°C.
PCR
Investigator: Sophie
PCR
| Name:
| Date:
|
| Continue from Experiment: new
|
| Project Name:
more NOT-gate with mutated restriction site (NHE I instead of Spe I)
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
| P 24 (1:10)
|
| 2.5µl
| Primer dw
| P 79 (1:10)
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
|
M45 (Bba_Q04400)
|
| 0.5 µl
| Phusion (add in the end)
|
|
The program we used: RT 2step (see last PCR of NOT-gate)

red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
PCR
| Name:Rüdiger
| Date: 29.08
|
| Continue from Experiment PCR Rüdiger (Date)-22.08.
(Name)
|
| Project Name: Precipitator
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
| P68Primer
|
| 2.5µl
| Primer dw
| P70Primer
|
| 1µl
| dNTPs
| of Template DNAPrimer
|
| 1µl
| DNA-Template
| GST-6-P-1 a,b,c,d,ePrimer
|
| 0.5 µl
| Phusion (add in the end)
|
|
What program do you use?
Annealing 60-70
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
| Name
| #
| Template
| Primer
| Anneal. Temp.
|
| For David Vector
Bam H1 +EcoR1
| 1
| Precipitator 3
| 87+88
| 65
|
| 2
| Precipitator 4_2
| 91+92
| 65
|
| 3
| Precipitator 4
| 98+90
| 65
|
| Gibson
| 4
| pGEX-6-p-1
| 85+44
| 66
|
| 5
| pGEX-6-p-1
| 85+45
| 66
|
| 6
| pGEX-6-p-1
| 85+46
| 66
|
| Registry cloning
| 7
| pGEX-6-p-1
| 85+48
| 66
|
| 8
| pGEX-6-p-1
| 48+84
| 66
|
| Toolbox Gibson>GST-6-p-1
| 9
| Precipitator 2
| 81+73
| 67
|
| 10
| Precipitator 4_2
| 82+71
| 67
|
| 11
| Precipitator 4
| 83+72
| 67
|
| Control
| 12
| Lambda DNA
| Primers
|
|
Testdigest of Precipitator in iGEM Vektor pSB1C3
Investigators: Julia and others
results see above under "green light receptor"
 "
"
 Contact
Contact 














