Team:Paris Bettencourt/Experiments/tRNA diffusion
From 2011.igem.org
Danyel.lee90 (Talk | contribs) (→Characterization of the tRNA biobrick) |
Danyel.lee90 (Talk | contribs) (→Characterization of the tRNA biobrick) |
||
| Line 5: | Line 5: | ||
In order to characterize the tRNA amber suppressor, as well as the pHyperspank, a special part was made: Pveg-SpoVG-GFP amber-TT. | In order to characterize the tRNA amber suppressor, as well as the pHyperspank, a special part was made: Pveg-SpoVG-GFP amber-TT. | ||
A double transformation was done with minipreps of two plasmids of different antibiotic resistances : pSB1C3 holding the tRNA under the control of pHyperspank and pSB1AK3 holding the GFP with the amber codon under the control of a constitutive promotor. Two groups of cultures of each picked colony were made. One group was cultured in rich medium without IPTG whilst the other group was cultured in rich medium with IPTG (4mM in 10mL). A culture of the strain holding GFP amber alone was grown in parallel. The two tubes of each colony were compared along with the negative control (GFP amber alone). Cultures were then diluted for microscopy, glycerols, and future experiments. We used the following strains to characterize this system: | A double transformation was done with minipreps of two plasmids of different antibiotic resistances : pSB1C3 holding the tRNA under the control of pHyperspank and pSB1AK3 holding the GFP with the amber codon under the control of a constitutive promotor. Two groups of cultures of each picked colony were made. One group was cultured in rich medium without IPTG whilst the other group was cultured in rich medium with IPTG (4mM in 10mL). A culture of the strain holding GFP amber alone was grown in parallel. The two tubes of each colony were compared along with the negative control (GFP amber alone). Cultures were then diluted for microscopy, glycerols, and future experiments. We used the following strains to characterize this system: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Direct observation under fluorescence lamp and microscopy was used to characterize this system. | Direct observation under fluorescence lamp and microscopy was used to characterize this system. | ||
Revision as of 00:21, 22 September 2011

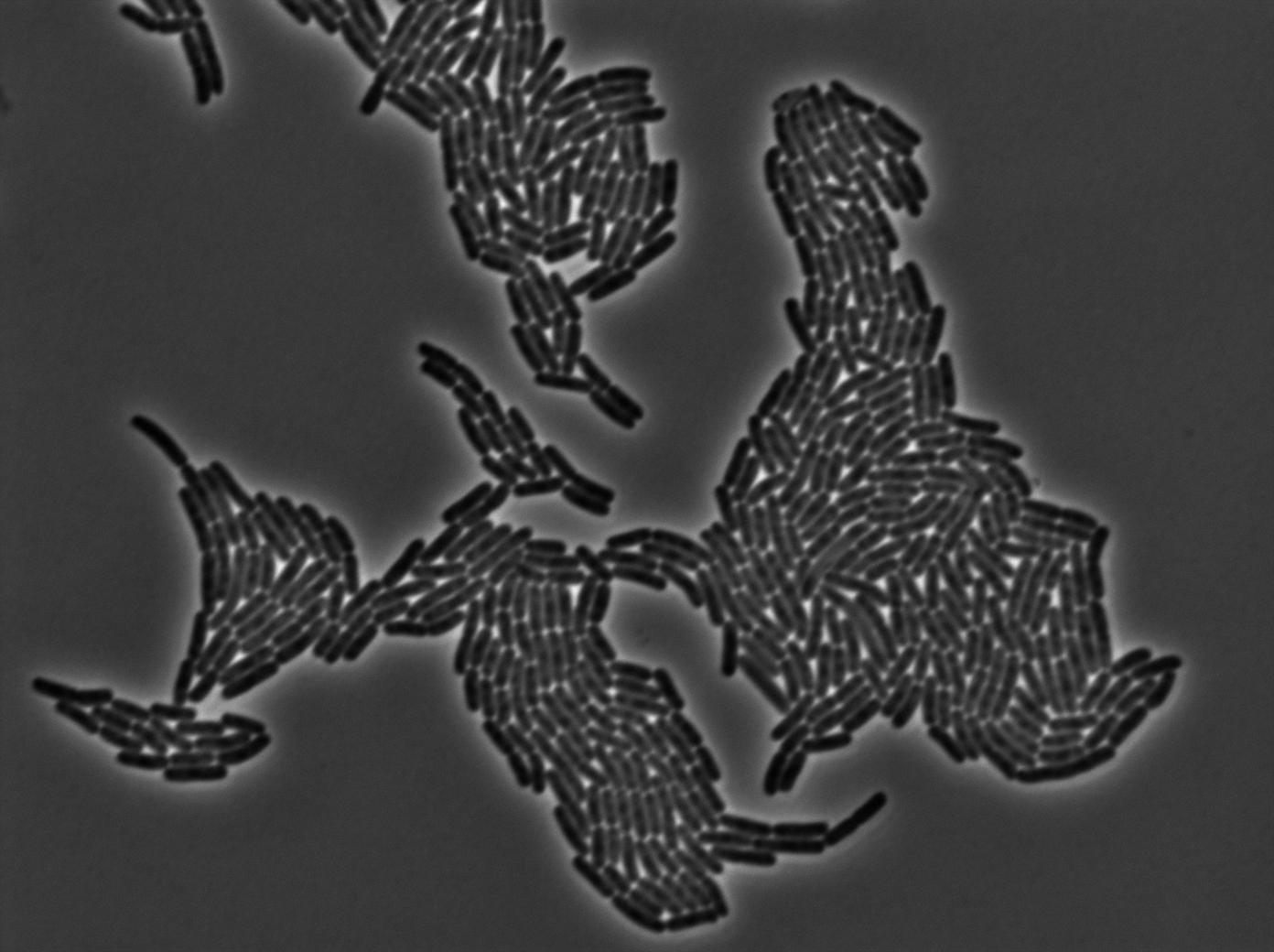
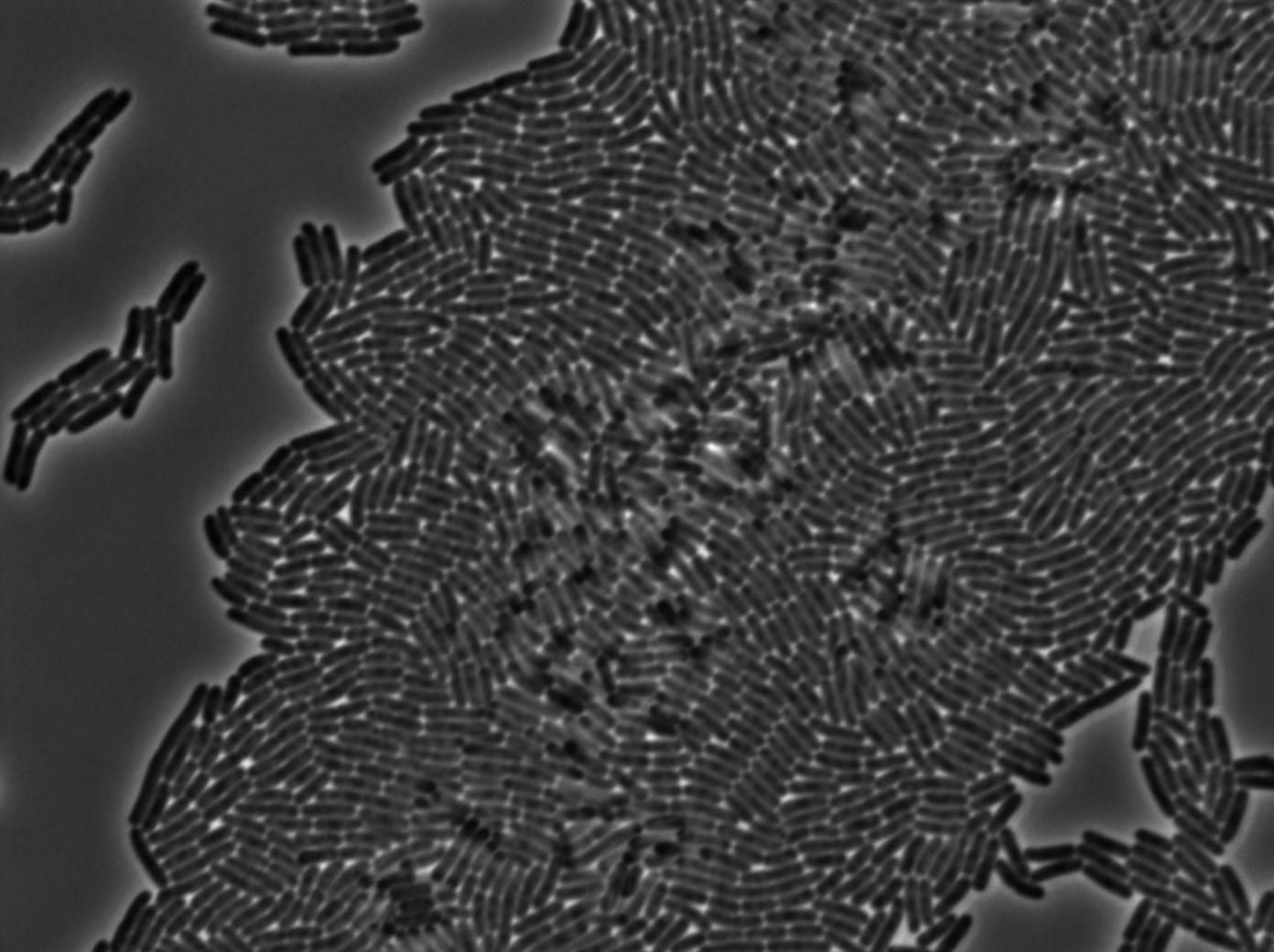
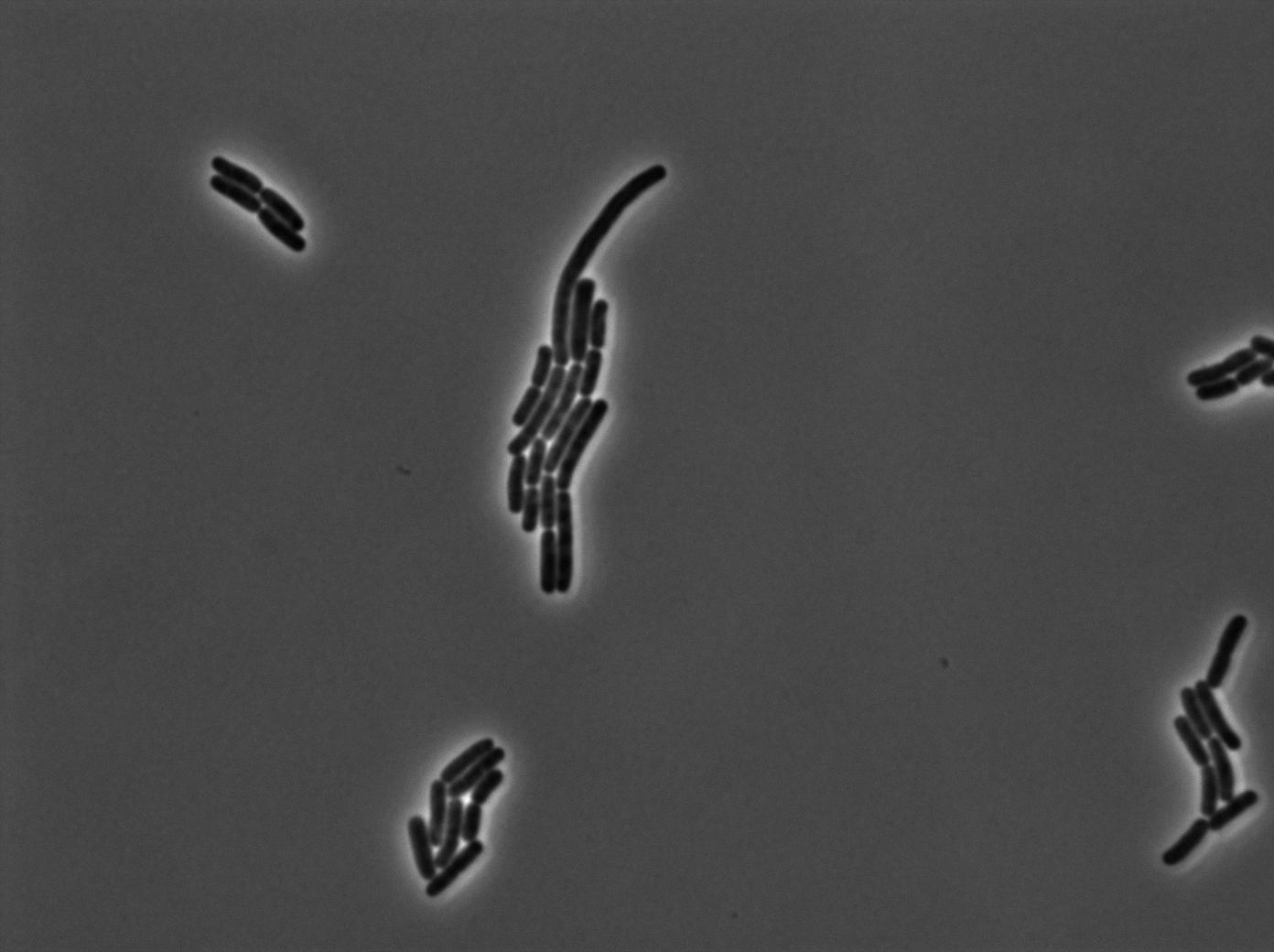
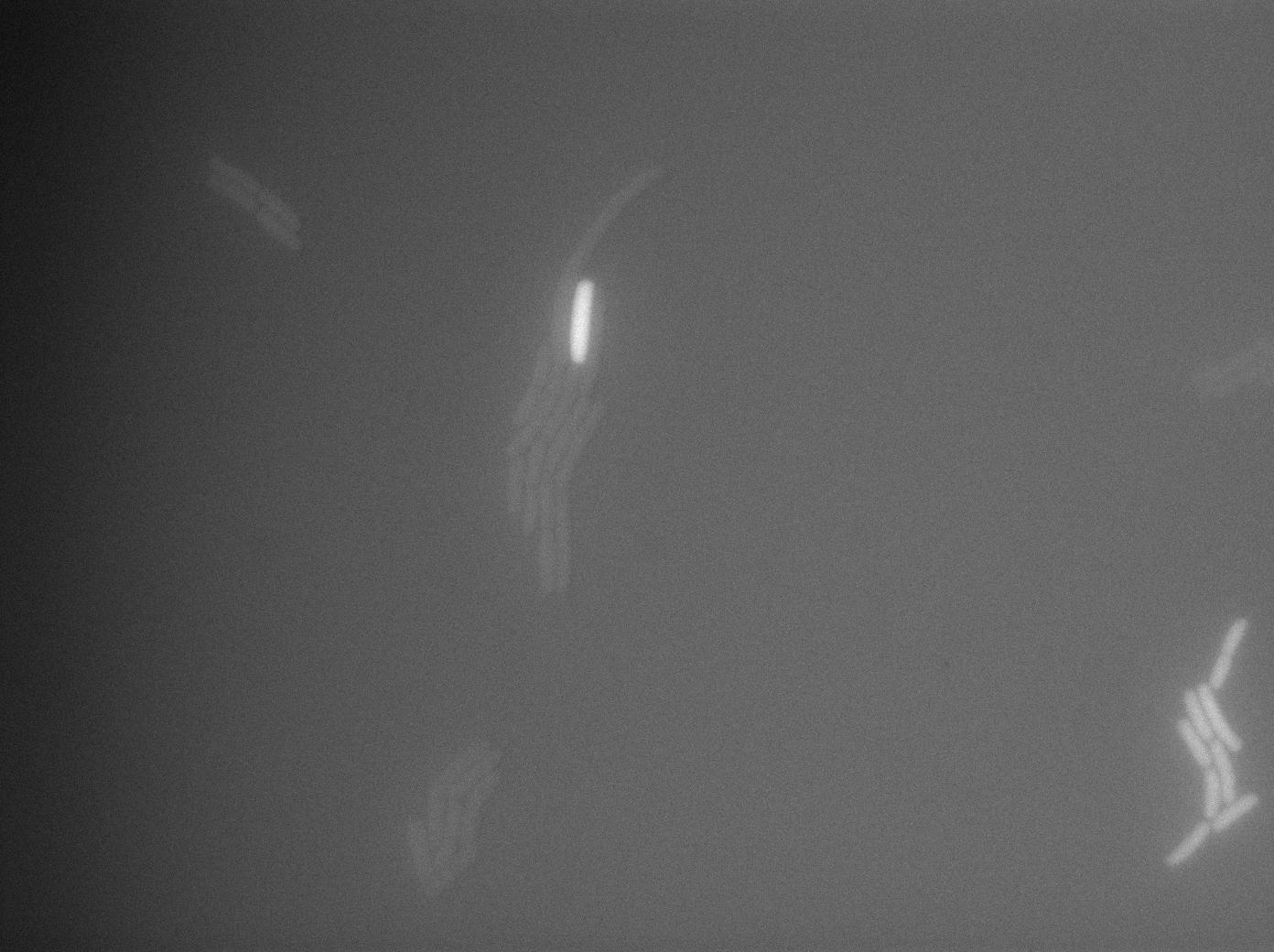
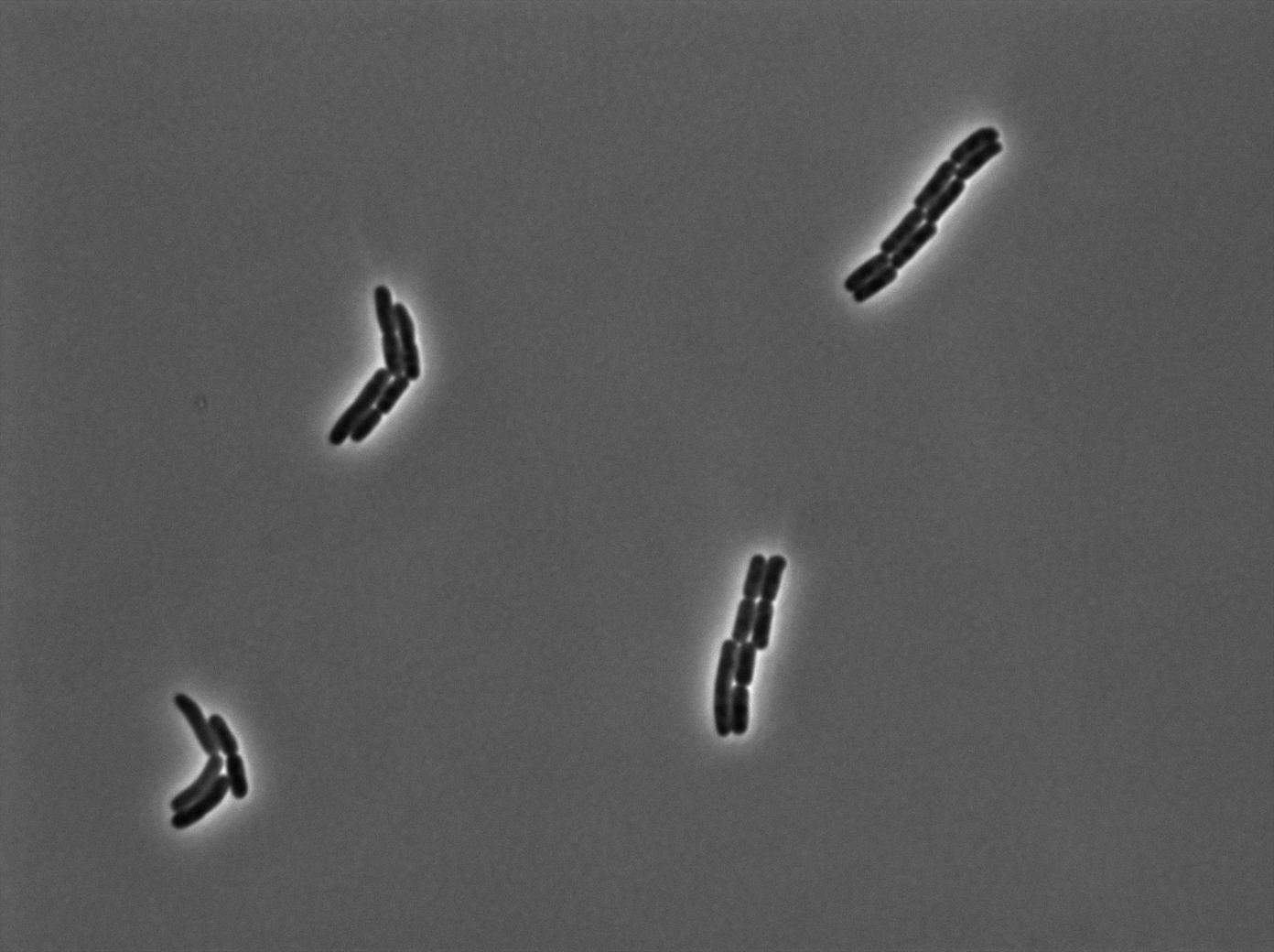
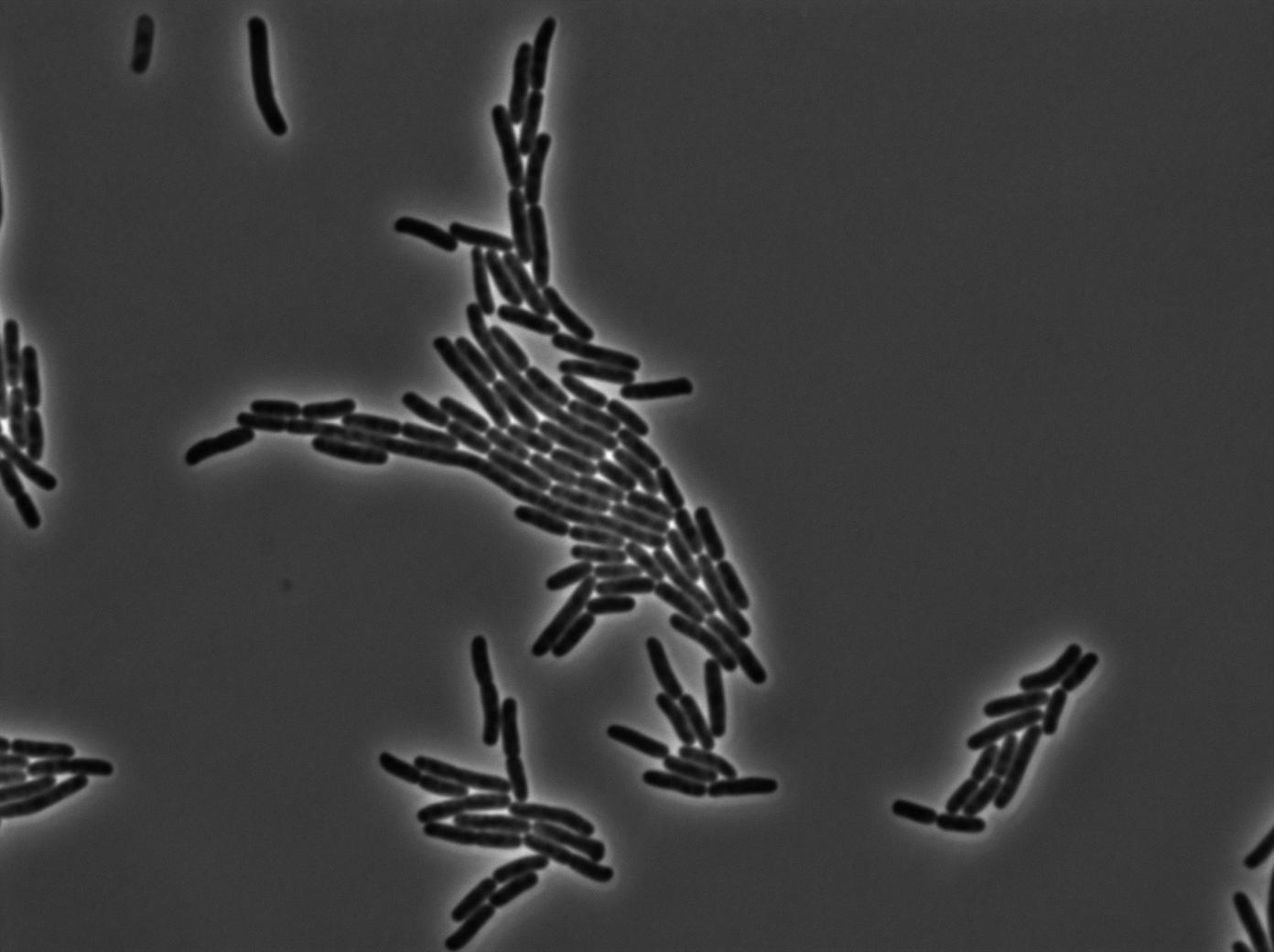
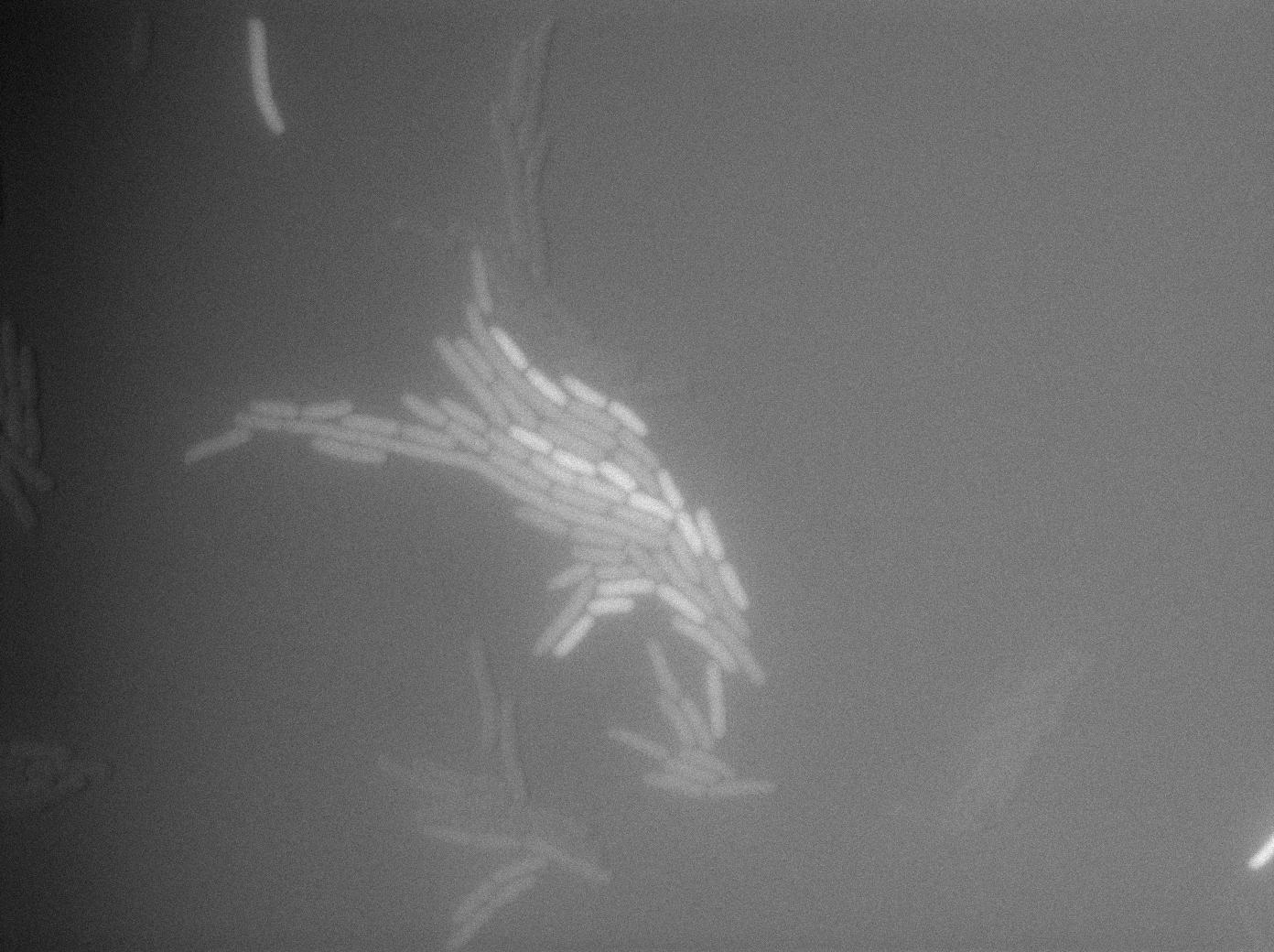
Characterization of the tRNA biobrick
In order to characterize the tRNA amber suppressor, as well as the pHyperspank, a special part was made: Pveg-SpoVG-GFP amber-TT. A double transformation was done with minipreps of two plasmids of different antibiotic resistances : pSB1C3 holding the tRNA under the control of pHyperspank and pSB1AK3 holding the GFP with the amber codon under the control of a constitutive promotor. Two groups of cultures of each picked colony were made. One group was cultured in rich medium without IPTG whilst the other group was cultured in rich medium with IPTG (4mM in 10mL). A culture of the strain holding GFP amber alone was grown in parallel. The two tubes of each colony were compared along with the negative control (GFP amber alone). Cultures were then diluted for microscopy, glycerols, and future experiments. We used the following strains to characterize this system:
Direct observation under fluorescence lamp and microscopy was used to characterize this system.
Fluorescence lamp
The cultures of strains having transformed both tRNA amber suppressor and GFP amber were strongly fluorescent, while the culture of the GFP amber alone as well as the culture of the colony without IPTG induction showed basal fluorescence.
Microscopy
 "
"