Team:EPF-Lausanne/Tools/Microfluidics
From 2011.igem.org
(→Microfluidics in synthetic biology) |
|||
| Line 5: | Line 5: | ||
== Microfluidics in synthetic biology == | == Microfluidics in synthetic biology == | ||
| - | Microfluidics is a powerful tool for biological research, having a wide range of applications. | + | Microfluidics is a powerful tool for biological research, having a wide range of applications. Essentially any molecular biology technique can be adapted, including: |
* On-chip gene synthesis: protein expression from coding DNA | * On-chip gene synthesis: protein expression from coding DNA | ||
| - | * On-chip chemostat chambers: can be used to trace the | + | * On-chip chemostat chambers: can be used to trace the fate of a single bacterium or to grow bacteria/yeast |
* Protein-protein interaction screening: for example between SH3 domains or to detect antibody binding | * Protein-protein interaction screening: for example between SH3 domains or to detect antibody binding | ||
* DNA-protein interactions: to determine the binding affinity of a transcription factor or to discover new regulatory proteins | * DNA-protein interactions: to determine the binding affinity of a transcription factor or to discover new regulatory proteins | ||
Revision as of 18:24, 21 September 2011
Microfluidics
Microfluidics Main | How-To Part I | How-To Part II | TamagotchipTo characterise our transcription factors, we used a lot of microfluidics, a tool underused by the iGEM community. We were lucky to have experts under hand to help us get started, so in turn here is our effort to promote microfluidics within the iGEM community, and help future teams use it.
Microfluidics in synthetic biology
Microfluidics is a powerful tool for biological research, having a wide range of applications. Essentially any molecular biology technique can be adapted, including:
- On-chip gene synthesis: protein expression from coding DNA
- On-chip chemostat chambers: can be used to trace the fate of a single bacterium or to grow bacteria/yeast
- Protein-protein interaction screening: for example between SH3 domains or to detect antibody binding
- DNA-protein interactions: to determine the binding affinity of a transcription factor or to discover new regulatory proteins
These experiments share the same advantages when ported on-chip: reagent volumes are reduced, therefore minimising cost, and the tiny size of each reaction chamber allows massive parallelisation of the experiment, enabling high-throughput screening. More specifically, in vitro gene synthesis becomes affordable at this scale. Our MITOMI chip contains 768 wells, all visible in one frame, which allowed us to quantify affinity of hundreds of protein-DNA combinations in a single picture. The small channels also allow fine control over reaction conditions. Most soluble reagents can be used, including DNA, proteins, molecule libraries, and so on - allowing much creativity in experimental design.
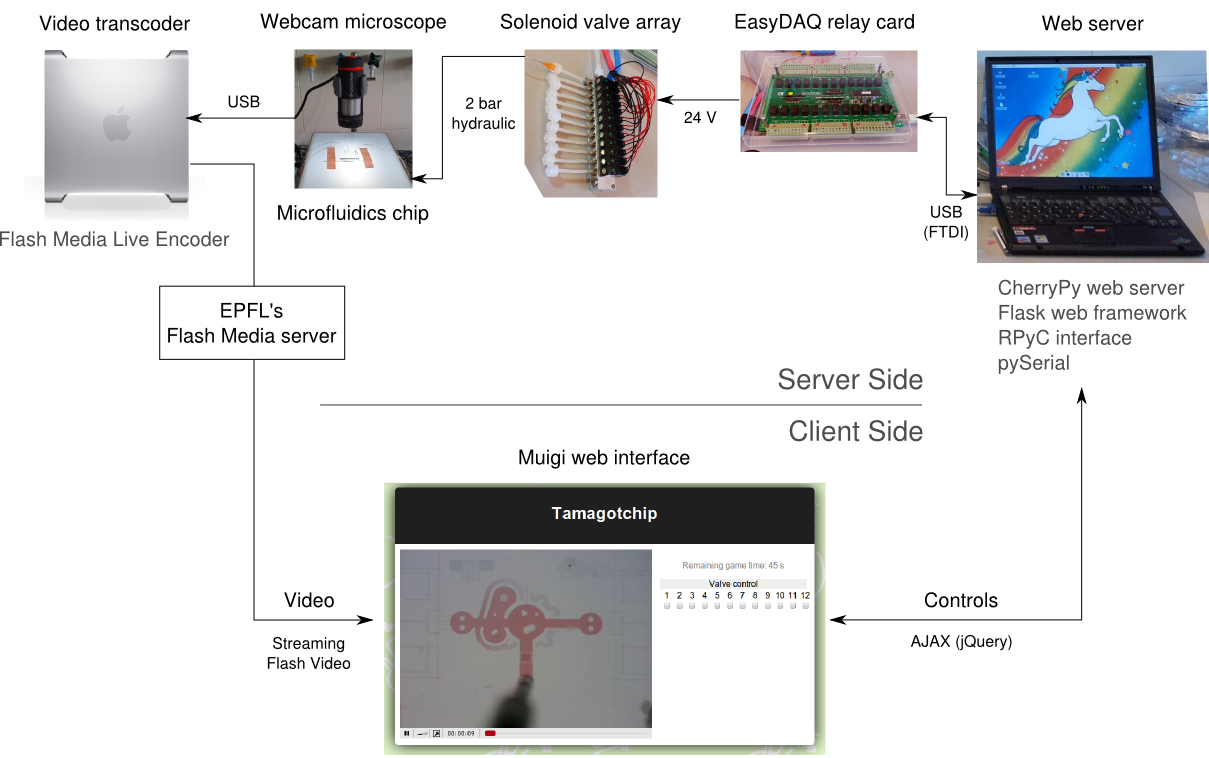
Few iGEM teams used this technology for their projects. We believe that promoting the use of microfluidics can help iGEM and improve the Parts Registry, specifically by providing more thorough part characterisation. To this end, we wrote a guide to get started with microfluidics, focusing on how to build a laboratory setup build a setup and outlining chip fabrication. To tickle the community's interest, we also created a live online microfluidics game, where iGEMers can control a chip located in our lab from their web browser.
 "
"