Team:Paris Bettencourt/Experiments/T7 diffusion
From 2011.igem.org
| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | |||
| + | <h1>T7 diffusion experiments</h1> | ||
<h2>Cloning</h2> | <h2>Cloning</h2> | ||
Revision as of 16:20, 21 September 2011

T7 diffusion experiments
Cloning
The cloning for this system, that seems simple at te begening took us a very long time, because of many issues we encounered along the cloning.
The first row of problems came from wrong parts sended by the registry. We lost almost one month and an halfin building the system. (see the corresponding page). We synthetized ourself the pHyperSpank promoter (K143055), but we didn't have the time to incorporate it into our biggest construction. Instead, we cloned a pVeg SpoVG promoter (K143053) that have a constitutive expression.
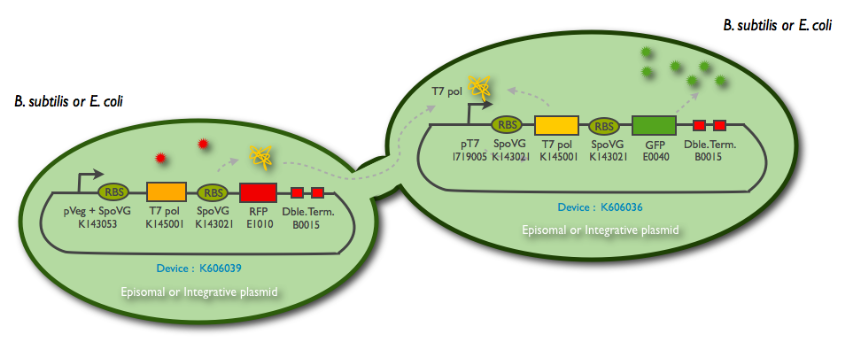
The following schematic represent what we have indeed managed to clone:
This summary shows the parts we have characterized, and the ones we have sended to the registry. But we have sucessfully managed to clone them into a integration and a replicative vector for subtilis, as well as the combination of the emittor an the receiver inside the same plasmid.
Diffusion experiments
We are around the corner of bringing this experiment under the microscope. We already have builded the microfluidic device, and it is just waiting for our cells.
At the moment, we have some difficulties with the transformation of our B. subtilis strain, but we hope we could show you great results and images for the Amsterdam iGEM Jamboree!!
Characterization of the T7 signal amplification leakage in E. Coli
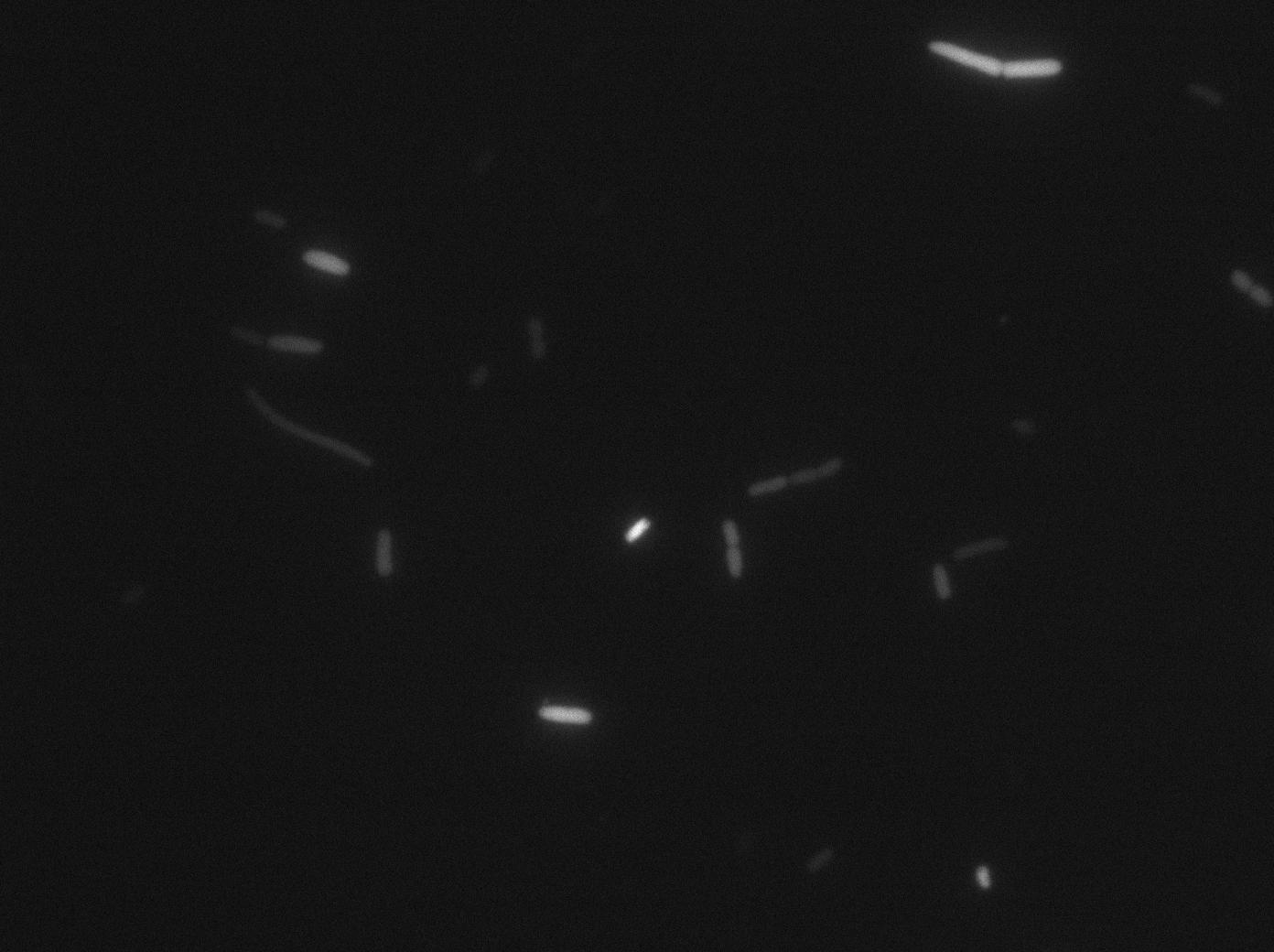
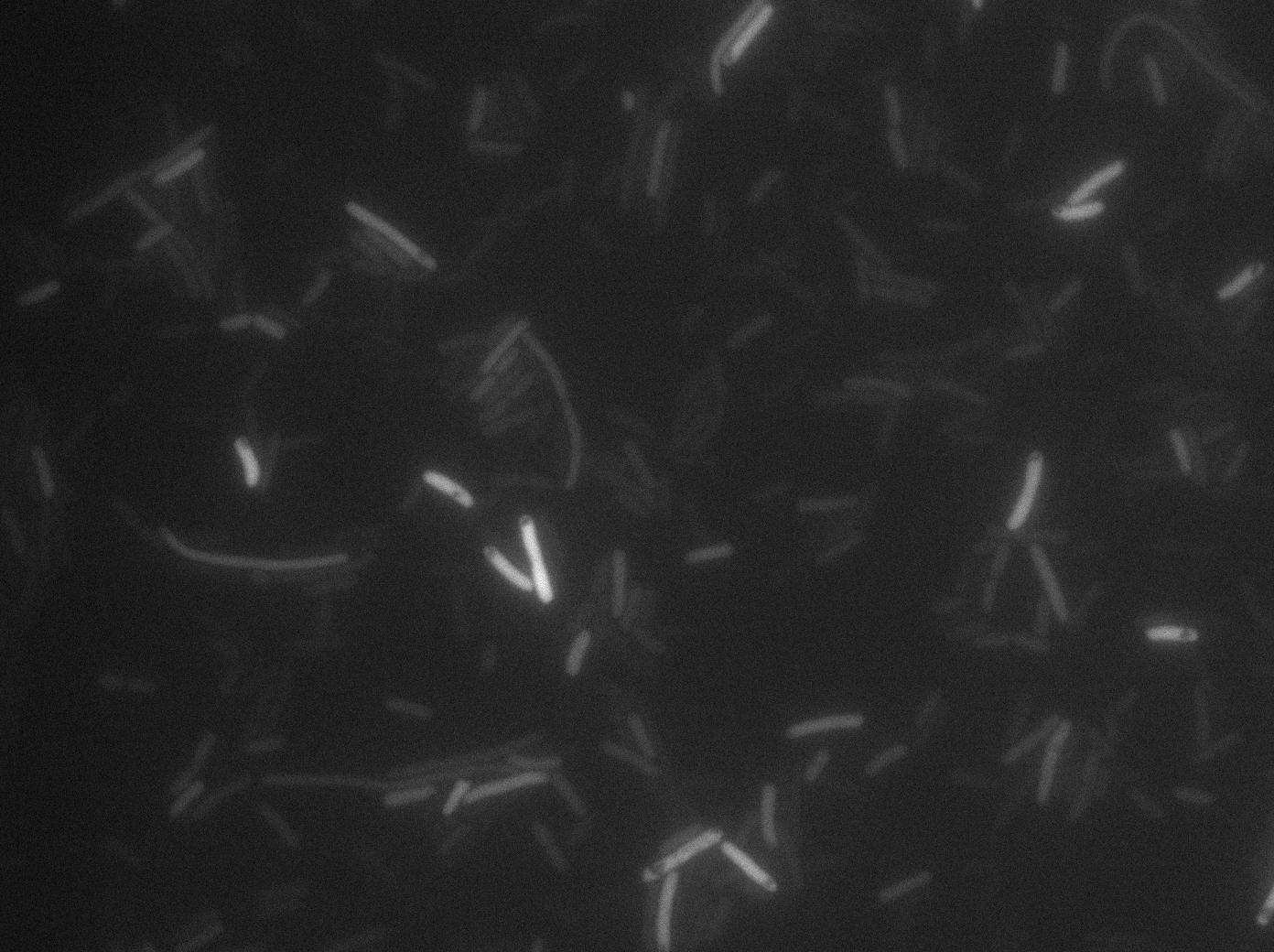
We characterized T7 autoloop in E. Coli, when hosted in the plasmid pSB1C3 (the K606036 part). The idea was to see if the system was leaky inside a synthetic biology plasmid, that is to say, a plasmid holding 4 terminators before any construct.
We found out that there is a leakage, but it is small. The cells in which the positive feed back look is activated stop deviding and glow with a very strong signal. Here are the images commented
The first pictures show that RFP construct has been well done. Indeed, we can see that some cells are glowing with RFP fluorescence. This also shows that the system is a bit leaky. Moreover the autoloop system is working very well because when leak occurs, cells glow highly.
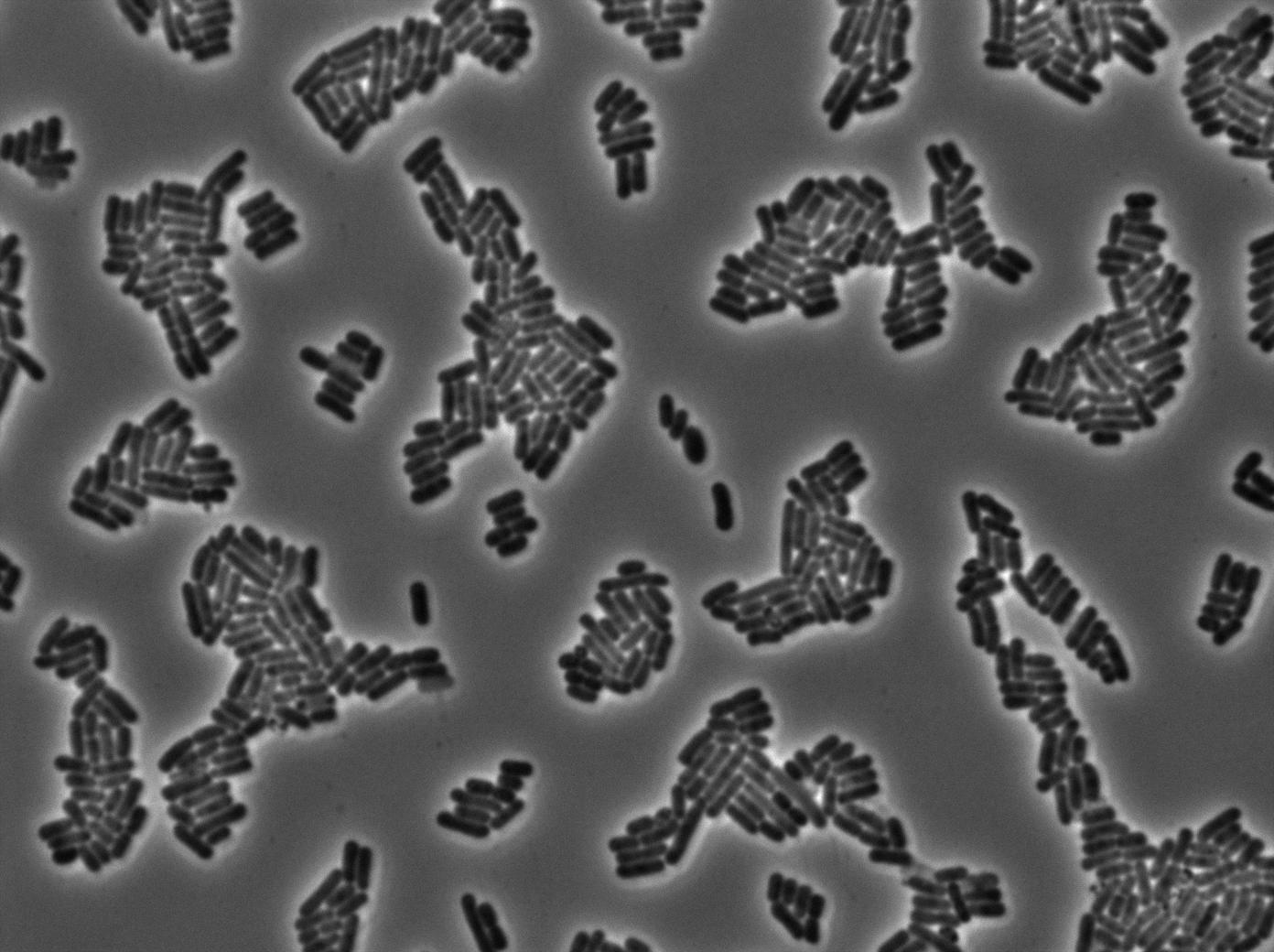
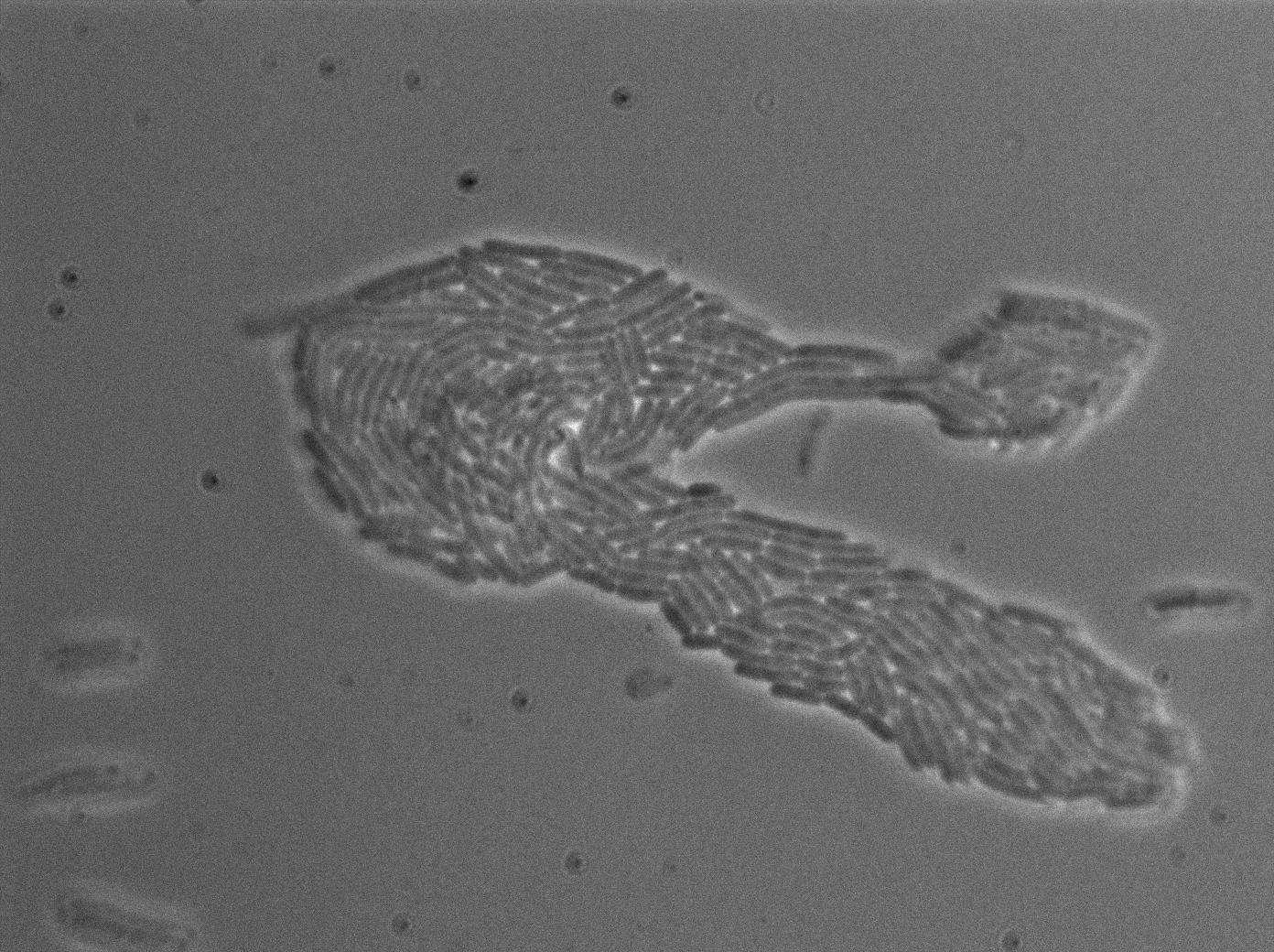
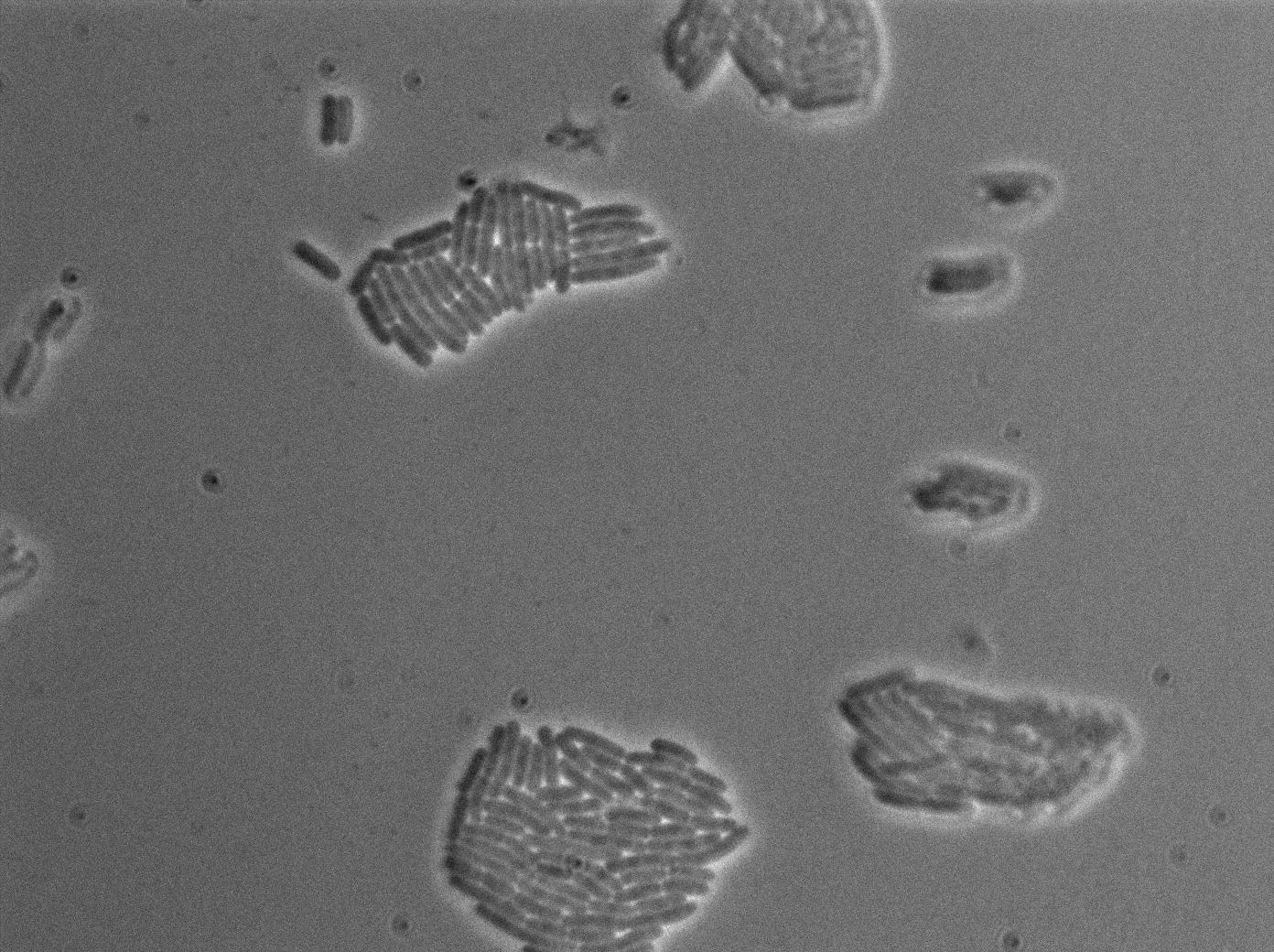
This pictures show that GFP system gives more disparity than the RFP construct. This is probably due to the LB that already glows in green. It is also noticeable that B.Subtilis is naturally shining green. Moreover, the absence of double terminator could explain the leakiness of our system. However, we expected this cells to glow in RFP but they did not. This shows that the full construct has failed.
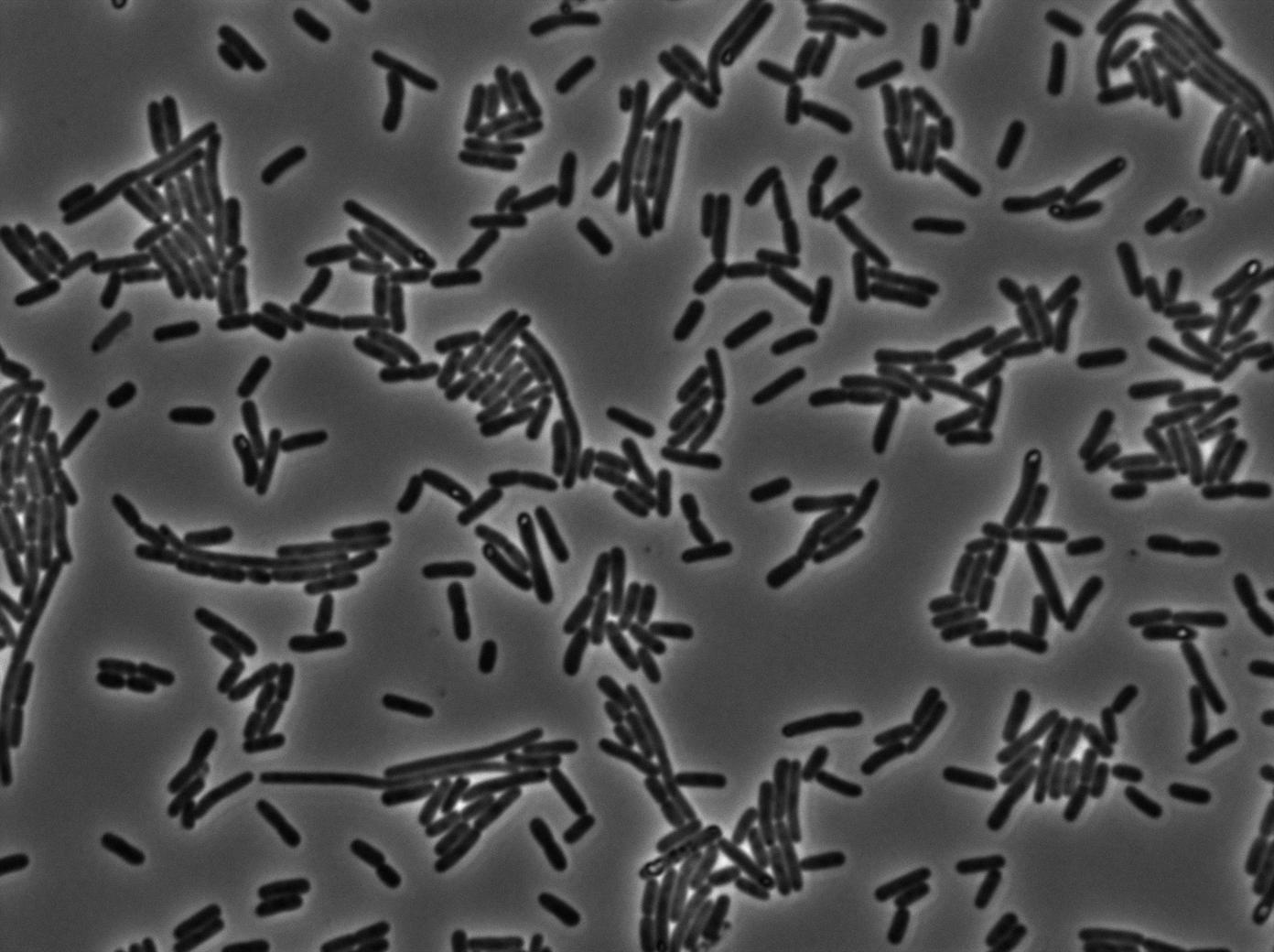
The last pictures show less disparity that the precedent one. This shows that double terminator actually proceeds on the phenotype. We also can notice that the cells glow in red a bit. However we can not say if it is an artifact or if the system is actually well constructed with the terminator and RFP construct.
 "
"