Team:DTU-Denmark/Background the natural system
From 2011.igem.org
(→The natural chitobiose system) |
(→The natural chitobiose system) |
||
| Line 5: | Line 5: | ||
[[File:DTU1_Chitobiose.png|350px|thumb|left|Chitobiose; a disaccharide derived from chitin.]] | [[File:DTU1_Chitobiose.png|350px|thumb|left|Chitobiose; a disaccharide derived from chitin.]] | ||
| - | The project is inspired by | + | The project is inspired by the regulation of chitobiose uptake and metabolism in ''E.coli''. Especially the following three players, which have indentified independently in several organisms and thus goes by many names: |
| + | |||
| + | ## a chitoporin that transports chitosugars into cells, called ''chiP'' (alias ''ybfM'') | ||
| + | |||
| + | ## a sRNA which regulates the chitoporin post-transcriptionally, called ''chiX'' (alias ''sroB'', ''micM'') | ||
| + | |||
| + | ## a trap-RNA which is transcribed from intergenic region in the chitobiose operon (chbBCARG), which we named ''chiXR'' for ''chiX'' regulator. | ||
| + | |||
| + | The sRNA regulates chitobiose (''chiP'') expression through binding to the Shine Dalgarno sequence on the ''chiP'' | ||
| + | mRNA inhibiting recruitment of the 30S ribosome and altering RNA stability. When chitobiose is present trap-RNA (''chiXR'') is transcribed and its transcript binds the sRNA (''chiX''), relieving chitoporin repression. | ||
| + | |||
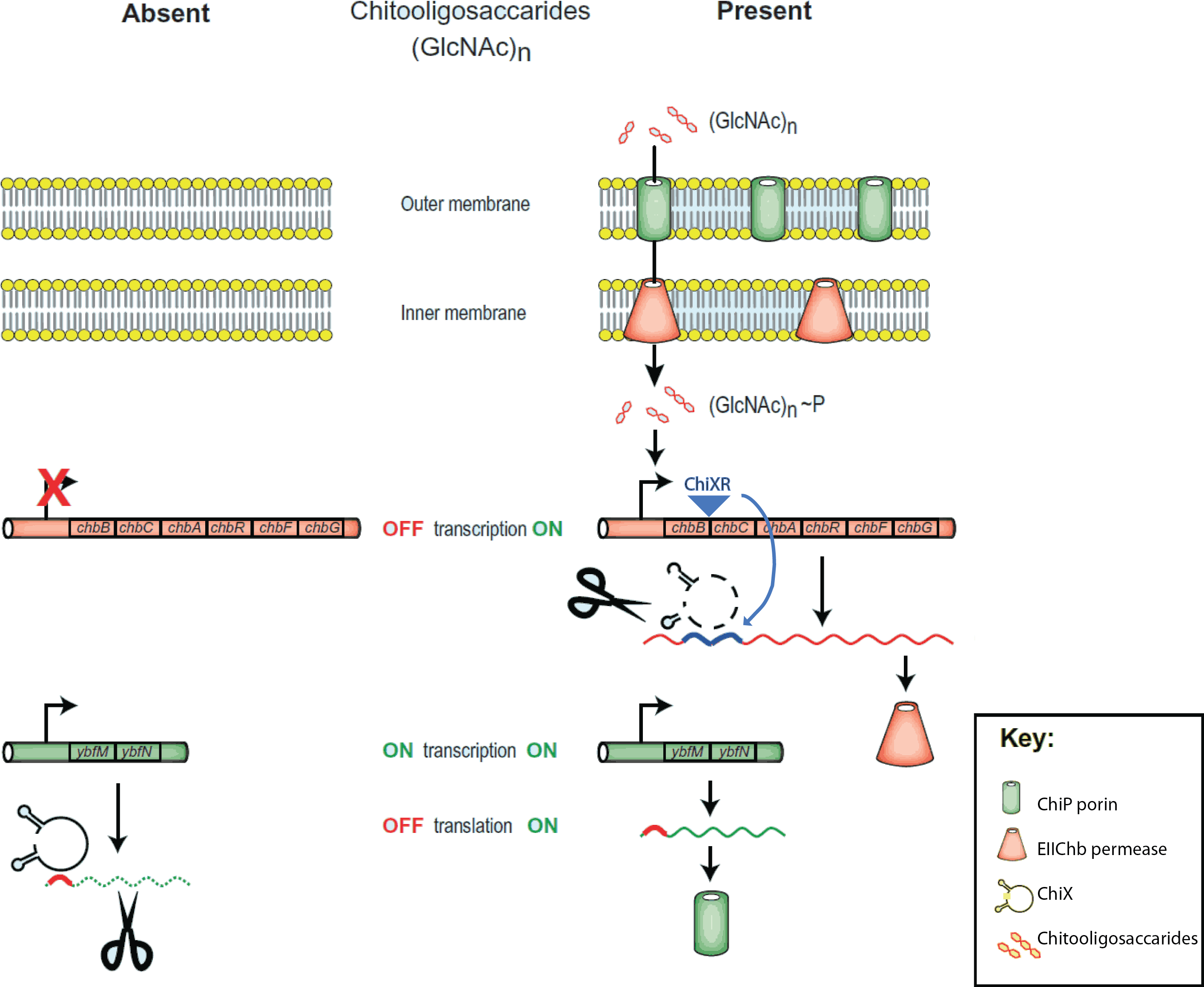
[[File:Chitobiose_system.png|350px|thumb|right|The ''E.coli'' chitobiose system. Figure 5 from<span class="superscript">[[#References|[2]]]</span>]] | [[File:Chitobiose_system.png|350px|thumb|right|The ''E.coli'' chitobiose system. Figure 5 from<span class="superscript">[[#References|[2]]]</span>]] | ||
Revision as of 13:14, 21 September 2011
Background
The natural chitobiose system
The project is inspired by the regulation of chitobiose uptake and metabolism in E.coli. Especially the following three players, which have indentified independently in several organisms and thus goes by many names:
- a chitoporin that transports chitosugars into cells, called chiP (alias ybfM)
- a sRNA which regulates the chitoporin post-transcriptionally, called chiX (alias sroB, micM)
- a trap-RNA which is transcribed from intergenic region in the chitobiose operon (chbBCARG), which we named chiXR for chiX regulator.
The sRNA regulates chitobiose (chiP) expression through binding to the Shine Dalgarno sequence on the chiP mRNA inhibiting recruitment of the 30S ribosome and altering RNA stability. When chitobiose is present trap-RNA (chiXR) is transcribed and its transcript binds the sRNA (chiX), relieving chitoporin repression.
References
[1] Figueroa-Bossi, Nara, Martina Valentini, Laurette Malleret, and Lionello Bossi. “Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target.” Genes & Development 23, no. 17 (2009): 2004 -2015. http://genesdev.cshlp.org/content/23/17/2004.abstract.
[2] Overgaard, Martin, Jesper Johansen, Jakob Møller‐Jensen, and Poul Valentin‐Hansen. “Switching off small RNA regulation with trap‐mRNA.” Molecular Microbiology 73, no. 5 (September 2009): 790-800. http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.2009.06807.x/abstract.
 "
"