Team:Groningen/project characterisation mu
From 2011.igem.org
(Difference between revisions)
| Line 4: | Line 4: | ||
==cI Autoinducing Loops== | ==cI Autoinducing Loops== | ||
The autoinducing loop driven by cI will function as our first memory unit. | The autoinducing loop driven by cI will function as our first memory unit. | ||
| - | We aimed to find a combination of RBS, transcription factor and degradation tag that enables us to toggle between the two steady states in a robust way. To estimate the rate in which single cells are activated by too much leakage of cI protein we measured the fluorescence in uninduced cells with a flow cytometer. To measure the activation we used a construct with the PRM promoter producing GFP. | + | We aimed to find a combination of RBS, transcription factor and degradation tag that enables us to toggle between the two steady states in a robust way. To estimate the rate in which single cells are activated by too much leakage of cI protein we measured the fluorescence in uninduced cells with a flow cytometer. To measure the activation we used a construct with the PRM promoter producing GFP with an LVA tag to make dynamic measurement possible. |
{| class="wikitable" | {| class="wikitable" | ||
| Line 10: | Line 10: | ||
|- | |- | ||
| [[File:ci1ui.png]] | | [[File:ci1ui.png]] | ||
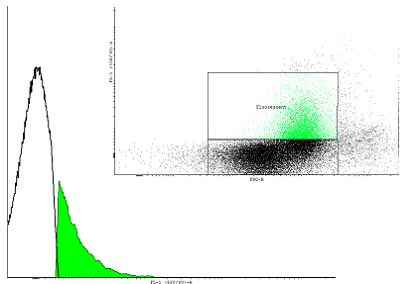
| + | Figure 1. A dotblot of uninduced cells carrying the pSB1K3 plasmid with the biobrick BBa_ and the plasmid pSB1A3 with as well an input(PBAD controlling cI) as well as the output (PRM controlling GFP-LVA). | ||
| | | | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 27: | Line 28: | ||
|} | |} | ||
|- | |- | ||
| - | | colspan=2 | Two populations are visible | + | | colspan=2 | Two populations are visible on the dotblot. We seperated both populations on the histogram |
|} | |} | ||
Revision as of 23:05, 20 September 2011
Memory Units
cI Autoinducing Loops
The autoinducing loop driven by cI will function as our first memory unit. We aimed to find a combination of RBS, transcription factor and degradation tag that enables us to toggle between the two steady states in a robust way. To estimate the rate in which single cells are activated by too much leakage of cI protein we measured the fluorescence in uninduced cells with a flow cytometer. To measure the activation we used a construct with the PRM promoter producing GFP with an LVA tag to make dynamic measurement possible.
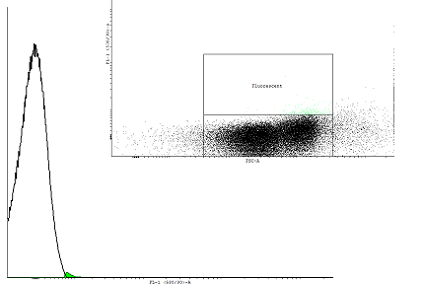
| cI autoinducing loop 6 (BBa_K00) after 4 hours of incubation | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| Only one population is visible blabla | |||||||||||||||||
 "
"