Team:Washington/Alkanes/Methods
From 2011.igem.org
(→Introducing the PetroBrick) |
|||
| Line 6: | Line 6: | ||
[[Image:Washington_2011_PetroBrick.png|220px|frameless|border="2"|link=http://partsregistry.org/wiki/index.php?title=Part:BBa_K590025|left]] | [[Image:Washington_2011_PetroBrick.png|220px|frameless|border="2"|link=http://partsregistry.org/wiki/index.php?title=Part:BBa_K590025|left]] | ||
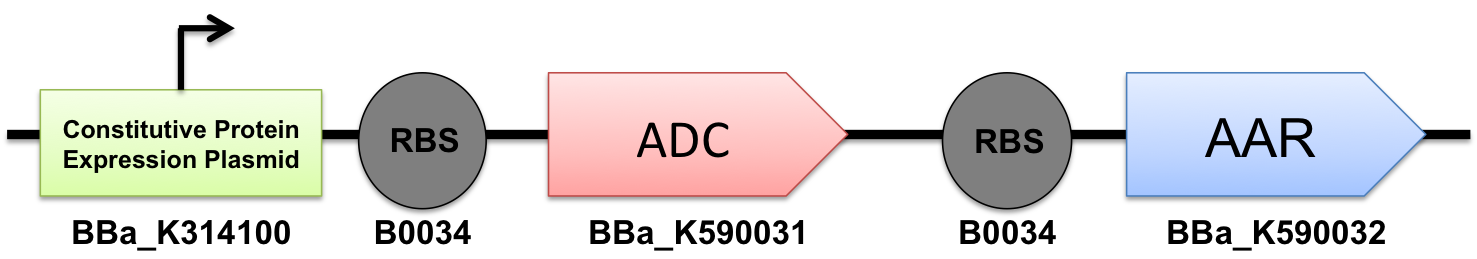
| - | In order to produce alkanes, we need both acyl-ACP reductase ([http://partsregistry.org/wiki/index.php?title=Part: | + | In order to produce alkanes, we need both [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 Part:BBa_K590031] acyl-ACP reductase ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 ADC]) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 Part:BBa_K590031] aldehyde decarbonylase ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 AAR]) to work together in the cell. In order to achieve this goal, we used standard cloning methods combining both to construct the [http://partsregistry.org/Part:BBa_K590025 BBa_K590025] Biobrick that contained both AAR and ADC under a high constitutive promoter, each with its own Elowitz standard RBS. This construct successfully synthesized our target product, and thus we have created a new modular alkane-producing platform: |
<br> | <br> | ||
| Line 18: | Line 18: | ||
---- | ---- | ||
| - | |||
=Alkane Production & Extraction= | =Alkane Production & Extraction= | ||
Revision as of 21:18, 20 September 2011
Introducing the PetroBrick
In order to produce alkanes, we need both [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 Part:BBa_K590031] acyl-ACP reductase ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 ADC]) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 Part:BBa_K590031] aldehyde decarbonylase ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 AAR]) to work together in the cell. In order to achieve this goal, we used standard cloning methods combining both to construct the [http://partsregistry.org/Part:BBa_K590025 BBa_K590025] Biobrick that contained both AAR and ADC under a high constitutive promoter, each with its own Elowitz standard RBS. This construct successfully synthesized our target product, and thus we have created a new modular alkane-producing platform:
the PetroBrick.
Alkane Production & Extraction
After we had the complete gene assembly in our hands, the next step was to transform it into cells and start them growing for alkane production. We let them grow in 37 degree shaker for 48-72 hours, in sealed glass tubes. After the cells have gone through the alkane production process, the next step is to extract the alkanes out of the cell broth. We add acyl acetate directly into the glass test tube for cell growth. Then we vortex until to everything is well mixed, to make sure all of the alkanes go directly into the ethyl acetate solvent. Next, we spin down the mixture by using a centrifuge at full speed to form three layers (cell pellet, media, and ethyl acetate supernatant). We use only the ethyl acetate layer to send for GCMS analysis.
Alkane Detection
We utilized a Gas Chromatograph / Mass Spectrometer (GCMS) to analyze alkane production concentrations. The GCMS is considered a "specific" test, because it identifies compounds specifically, not just to a category of compounds. It works by separating the individual components of a sample through a capillary column based mainly on it's boiling point, similar to fractional distillation. The separated compounds generally elute from the column at different retention times, and are passed to the mass spectrometer. Each compound is then broken down into it's individual molecular components through electron stream ionization. These ions differ in mass-to-charge (m/Q) ratios, creating a unique ion de-composition profile for each compound that can be used to identify it through comparison to known chemical standards. Because compounds occasionally have similar elution times or mass spec fingerprints, the combination of analyses results in reducing the chances for overlap.
 "
"