Team:EPF-Lausanne/Our Project/Data
From 2011.igem.org
(→Readout system - TetR and lysis) |
(→Readout system - TetR, LacI and RFP) |
||
| Line 53: | Line 53: | ||
To get this readout system, we cotransformed pSB3K1 Pconst-TetR Ptet-LacI with J61002 Plac-RFP. Unfortunately, the sequence of Ptet in front of LacI got mutated during the assembly process, resulting in only a partial repression of LacI by TetR. Still, our results do show the effects of TetR and LacI on the whole system. | To get this readout system, we cotransformed pSB3K1 Pconst-TetR Ptet-LacI with J61002 Plac-RFP. Unfortunately, the sequence of Ptet in front of LacI got mutated during the assembly process, resulting in only a partial repression of LacI by TetR. Still, our results do show the effects of TetR and LacI on the whole system. | ||
| + | [[File:EPFL_Summary_TetR_LacI_RFP.jpg]] | ||
''ATC induction'' | ''ATC induction'' | ||
Revision as of 18:19, 17 September 2011
Data
That's how the Data page should look like: https://igem.org/Sample_Data_Page Perhaps we could only provide the link to the Registry page; if we put all the graphs here it's gonna be messy pages (TetR mutants, reporter plasmids and T7 variants) We can make a "results" page with all the graphs, or put them in the descript
Contents |
New parts
TetR Mutants
Lilia, are you also putting the WT into biobrick?
- V36F
Sequence
ATGTCCAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGCTGCTTAATGAGGTCGGAATCGAAGGTTTAACAACCCGTAAACTCGCCCAGAAGCTAGGTTTCGAGCAGCCTACATTGTATTGGCATGTAAAAAATAAGCGGGCTTTGCTCGACGCCTTAGCCATTGAGATGTTAGATAGGCACCATACTCACTTTTGCCCTTTAGAAGGGGAAAGCTGGCAAGATTTTTTACGTAATAACGCTAAAAGTTTTAGATGTGCTTTACTAAGTCATCGCGATGGAGCAAAAGTACATTTAGGTACACGGCCTACAGAAAAACAGTATGAAACTCTCGAAAATCAATTAGCCTTTTTATGCCAACAAGGTTTTTCACTAGAGAATGCATTATATGCACTCAGCGCTGTGGGGCATTTTACTTTAGGTTGCGTATTGGAAGATCAAGAGCATCAAGTCGCTAAAGAAGAAAGGGAAACACCTACTACTGATAGTATGCCGCCATTATTACGACAAGCTATCGAATTATTTGATCACCAAGGTGCAGAGCCAGCCTTCTTATTCGGCCTTGAATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCT
- P39K
- Y42F
- P39Q-Y42M
Sequence
ATGTCCAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGCTGCTTAATGAGGTCGGAATCGAAGGTTTAACAACCCGTAAACTCGCCCAGAAGCTAGGTGTAGAGCAGCAAACAGTGATGTGGCATGTAAAAAATAAGCGGGCTTTGCTCGACGCCTTAGCCATTGAGATGTTAGATAGGCACCATACTCACTTTTGCCCTTTAGAAGGGGAAAGCTGGCAAGATTTTTTACGTAATAACGCTAAAAGTTTTAGATGTGCTTTACTAAGTCATCGCGATGGAGCAAAAGTACATTTAGGTACACGGCCTACAGAAAAACAGTATGAAACTCTCGAAAATCAATTAGCCTTTTTATGCCAACAAGGTTTTTCACTAGAGAATGCATTATATGCACTCAGCGCTGTGGGGCATTTTACTTTAGGTTGCGTATTGGAAGATCAAGAGCATCAAGTCGCTAAAGAAGAAAGGGAAACACCTACTACTGATAGTATGCCGCCATTATTACGACAAGCTATCGAATTATTTGATCACCAAGGTGCAGAGCCAGCCTTCTTATTCGGCCTTGAATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCT
T7 Promoter Variants
Other assemblies - without new biobrick
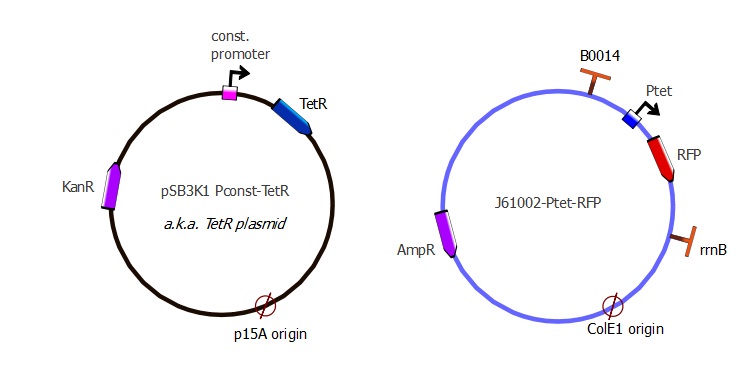
J61002 Ptet-RFP
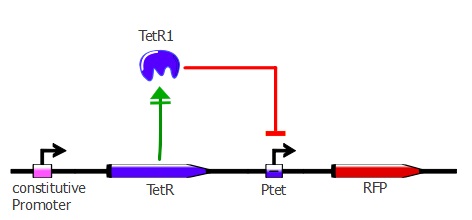
Readout system - TetR and RFP
The first readout system is composed of TetR with a constitutive promoter, followed by RFP with a pTet promoter. If TetR binds to pTet, then RFP is repressed. The plasmids used here are pSB3K1 pConst-TetR and J61002 Ptet-RFP. This readout system is convenient for fluorescence detection experiments, but it would not be suited for using the lysis cassette as the reporter gene is being repressed upon TetR-pTet interaction. With the lysis device, the interesting cells (where TetR binds to pTet) would survive and we would recover only the useless TetR mutants.
ATC induction
Dose-response
J61002 only
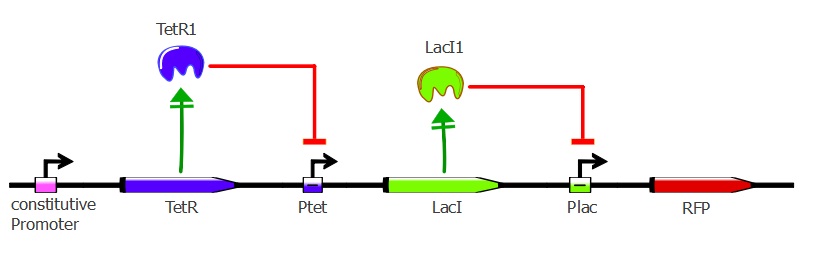
Readout system - TetR, LacI and RFP
This second readout system is composed of 3 genes: TetR under pConst control, LacI under pTet control (playing the role of an inverter) and finally RFP under pLac control. Here, RFP is induced when TetR binds to pTet. Lysis cassette can be put instead of RFP in thy system, having as a consequence that the cells where TetR mutants bind to pTet would lyse.
To get this readout system, we cotransformed pSB3K1 Pconst-TetR Ptet-LacI with J61002 Plac-RFP. Unfortunately, the sequence of Ptet in front of LacI got mutated during the assembly process, resulting in only a partial repression of LacI by TetR. Still, our results do show the effects of TetR and LacI on the whole system.
ATC induction
ATC dose-response curve
IPTG induction
IPTG dose-response curve
 "
"