Team:EPF-Lausanne/Our Project/Data
From 2011.igem.org
(→T7 Promoter Variants) |
|||
| Line 33: | Line 33: | ||
| - | === | + | === Readout system - TetR and RFP === |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | The first readout system is composed of TetR with a constitutive promoter, followed by RFP with a pTet promoter. If TetR '''binds''' to pTet, then RFP is ''' repressed'''. The plasmids used here are pSB3K1 pConst-TetR and J61002 Ptet-RFP. This readout system is convenient for fluorescence detection experiments, but it would not be suited for using the lysis cassette as the reporter gene is being repressed upon TetR-pTet interaction. | ||
''Sequence'' | ''Sequence'' | ||
| Line 65: | Line 55: | ||
''ATC induction'' | ''ATC induction'' | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
''Dose-response'' | ''Dose-response'' | ||
| - | |||
| - | |||
| - | |||
=== pSB3K1 Pconst-TetR Ptet-LacI === | === pSB3K1 Pconst-TetR Ptet-LacI === | ||
Revision as of 17:48, 17 September 2011
Data
That's how the Data page should look like: https://igem.org/Sample_Data_Page Perhaps we could only provide the link to the Registry page; if we put all the graphs here it's gonna be messy pages (TetR mutants, reporter plasmids and T7 variants) We can make a "results" page with all the graphs, or put them in the descript
Contents |
New parts
TetR Mutants
Lilia, are you also putting the WT into biobrick?
- V36F
Sequence
ATGTCCAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGCTGCTTAATGAGGTCGGAATCGAAGGTTTAACAACCCGTAAACTCGCCCAGAAGCTAGGTTTCGAGCAGCCTACATTGTATTGGCATGTAAAAAATAAGCGGGCTTTGCTCGACGCCTTAGCCATTGAGATGTTAGATAGGCACCATACTCACTTTTGCCCTTTAGAAGGGGAAAGCTGGCAAGATTTTTTACGTAATAACGCTAAAAGTTTTAGATGTGCTTTACTAAGTCATCGCGATGGAGCAAAAGTACATTTAGGTACACGGCCTACAGAAAAACAGTATGAAACTCTCGAAAATCAATTAGCCTTTTTATGCCAACAAGGTTTTTCACTAGAGAATGCATTATATGCACTCAGCGCTGTGGGGCATTTTACTTTAGGTTGCGTATTGGAAGATCAAGAGCATCAAGTCGCTAAAGAAGAAAGGGAAACACCTACTACTGATAGTATGCCGCCATTATTACGACAAGCTATCGAATTATTTGATCACCAAGGTGCAGAGCCAGCCTTCTTATTCGGCCTTGAATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCT
- P39K
- Y42F
- P39Q-Y42M
Sequence
ATGTCCAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGCTGCTTAATGAGGTCGGAATCGAAGGTTTAACAACCCGTAAACTCGCCCAGAAGCTAGGTGTAGAGCAGCAAACAGTGATGTGGCATGTAAAAAATAAGCGGGCTTTGCTCGACGCCTTAGCCATTGAGATGTTAGATAGGCACCATACTCACTTTTGCCCTTTAGAAGGGGAAAGCTGGCAAGATTTTTTACGTAATAACGCTAAAAGTTTTAGATGTGCTTTACTAAGTCATCGCGATGGAGCAAAAGTACATTTAGGTACACGGCCTACAGAAAAACAGTATGAAACTCTCGAAAATCAATTAGCCTTTTTATGCCAACAAGGTTTTTCACTAGAGAATGCATTATATGCACTCAGCGCTGTGGGGCATTTTACTTTAGGTTGCGTATTGGAAGATCAAGAGCATCAAGTCGCTAAAGAAGAAAGGGAAACACCTACTACTGATAGTATGCCGCCATTATTACGACAAGCTATCGAATTATTTGATCACCAAGGTGCAGAGCCAGCCTTCTTATTCGGCCTTGAATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCT
T7 Promoter Variants
Other assemblies - without new biobrick
J61002 Ptet-RFP
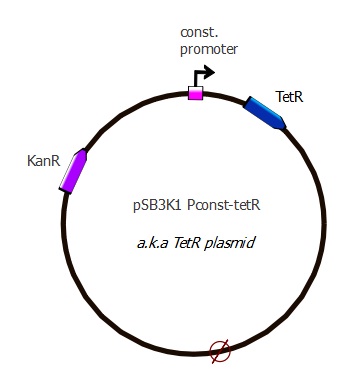
Readout system - TetR and RFP
The first readout system is composed of TetR with a constitutive promoter, followed by RFP with a pTet promoter. If TetR binds to pTet, then RFP is repressed. The plasmids used here are pSB3K1 pConst-TetR and J61002 Ptet-RFP. This readout system is convenient for fluorescence detection experiments, but it would not be suited for using the lysis cassette as the reporter gene is being repressed upon TetR-pTet interaction.
Sequence
Complete sequence of the plasmid: "pSb3K1 Pconst-TetR"
Do we really need this? Sequencing data compared to the sequence of Pconst,RBS+spacer and TetR gene:"pSb3K1_TetR_seq" All these parts are correct.
Plasmid map
This plasmid contains a p15A replication origin as well as a Kanamycin resistance marker.
ATC induction
Dose-response
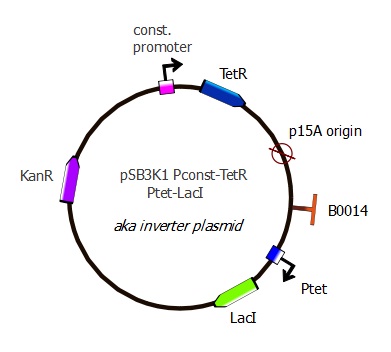
pSB3K1 Pconst-TetR Ptet-LacI
Description
This is the final TetR plasmid, containing the TetR gene as well as the LacI inverter with a Ptet promoter. Here we have wild-type TetR and Ptet sequences, but this plasmid is also intended to be used with mutant TetR genes and mutant Ptet binding sequences.
Parts assembled
- Plasmid backbone containing Pconst and TetR: see precedent section
- Terminator: B0014 from the Registry [http://partsregistry.org/Part:BBa_B0014 "B0014"] (sequence copied into our primers)
- Ptet: R0040 from Registry [http://partsregistry.org/Part:BBa_R0040 "R0040"] (sequence copied into our primers)
- LacI: amplified from Repressilator plasmid "LacI sequence" The sequence lacks a stop codon, we added TAA with our primers.
Sequence
Add results for TetR sequencing Add results for LacI asap
Plasmid map
This plasmid has the same structure as pSB3K1 Pconst-TetR: p15A replication origin and Kanamycin resistance marker. However, Ptet-LacI has been added after the p15A sequence.
ATC induction
ATC dose-response curve
IPTG induction
IPTG dose-response curve
 "
"