Team:EPF-Lausanne/Our Project/Data
From 2011.igem.org
m (→T7 Promoter Variants) |
(→T7 Promoter Variants) |
||
| Line 44: | Line 44: | ||
[[File:varability_comparison.png|200px]] | [[File:varability_comparison.png|200px]] | ||
| - | + | For the three sets of randomers for T7 with and without the lac operator, we tested seventy-two different variants and characterized their expression using the same IPTG induction protocol as with the designed variants. The goal of using these variants was to examine the range of expressions that can be produced by random mutations as opposed to directed mutations. The results, as presented in the graph, indicate that the designed variants (with and without lac operator put together) produce a much higher average normalized fluorescence than the randomers. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Other assemblies - without new biobrick == | == Other assemblies - without new biobrick == | ||
Revision as of 17:42, 16 September 2011
Data
That's how the Data page should look like: https://igem.org/Sample_Data_Page Perhaps we could only provide the link to the Registry page; if we put all the graphs here it's gonna be messy pages (TetR mutants, reporter plasmids and T7 variants) We can make a "results" page with all the graphs, or put them in the descript
Contents |
New parts
TetR Mutants
Lilia, are you also putting the WT into biobrick?
- V36F
Sequence
ATGTCCAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGCTGCTTAATGAGGTCGGAATCGAAGGTTTAACAACCCGTAAACTCGCCCAGAAGCTAGGTTTCGAGCAGCCTACATTGTATTGGCATGTAAAAAATAAGCGGGCTTTGCTCGACGCCTTAGCCATTGAGATGTTAGATAGGCACCATACTCACTTTTGCCCTTTAGAAGGGGAAAGCTGGCAAGATTTTTTACGTAATAACGCTAAAAGTTTTAGATGTGCTTTACTAAGTCATCGCGATGGAGCAAAAGTACATTTAGGTACACGGCCTACAGAAAAACAGTATGAAACTCTCGAAAATCAATTAGCCTTTTTATGCCAACAAGGTTTTTCACTAGAGAATGCATTATATGCACTCAGCGCTGTGGGGCATTTTACTTTAGGTTGCGTATTGGAAGATCAAGAGCATCAAGTCGCTAAAGAAGAAAGGGAAACACCTACTACTGATAGTATGCCGCCATTATTACGACAAGCTATCGAATTATTTGATCACCAAGGTGCAGAGCCAGCCTTCTTATTCGGCCTTGAATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCT
- P39K
- Y42F
- P39Q-Y42M
Sequence
ATGTCCAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGCTGCTTAATGAGGTCGGAATCGAAGGTTTAACAACCCGTAAACTCGCCCAGAAGCTAGGTGTAGAGCAGCAAACAGTGATGTGGCATGTAAAAAATAAGCGGGCTTTGCTCGACGCCTTAGCCATTGAGATGTTAGATAGGCACCATACTCACTTTTGCCCTTTAGAAGGGGAAAGCTGGCAAGATTTTTTACGTAATAACGCTAAAAGTTTTAGATGTGCTTTACTAAGTCATCGCGATGGAGCAAAAGTACATTTAGGTACACGGCCTACAGAAAAACAGTATGAAACTCTCGAAAATCAATTAGCCTTTTTATGCCAACAAGGTTTTTCACTAGAGAATGCATTATATGCACTCAGCGCTGTGGGGCATTTTACTTTAGGTTGCGTATTGGAAGATCAAGAGCATCAAGTCGCTAAAGAAGAAAGGGAAACACCTACTACTGATAGTATGCCGCCATTATTACGACAAGCTATCGAATTATTTGATCACCAAGGTGCAGAGCCAGCCTTCTTATTCGGCCTTGAATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCT
T7 Promoter Variants
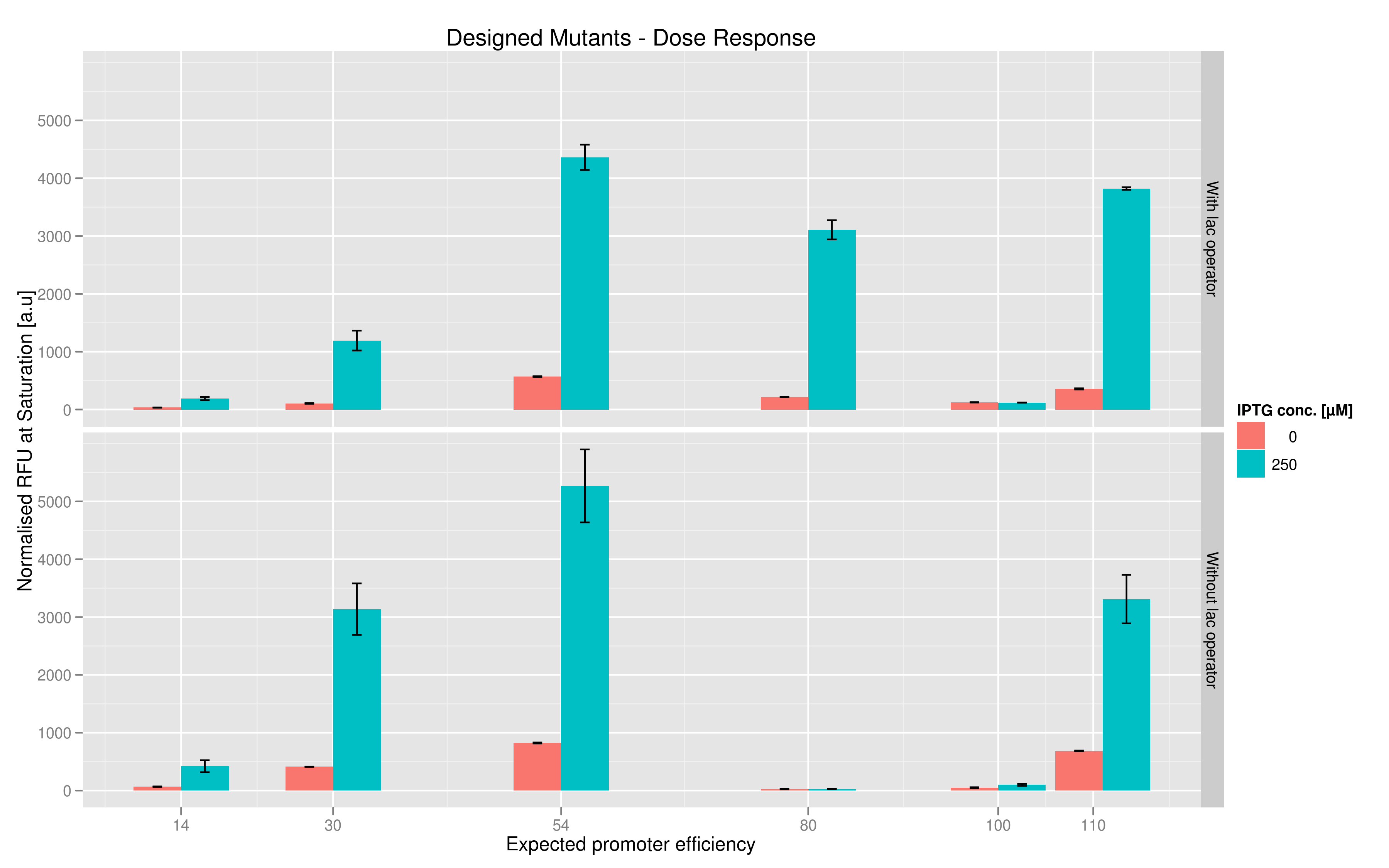
We made two families of T7 promoter variants. One family has mutations on the T7 promoter consensus sequence while the other has the same set of mutations on the consensus sequence but also has a lac operator downstream of the T7 promoter (but upstream of the reporter RFP or Lysis). In each family, we made six designed variants with different predicted promoter strengths compared to the wildtype and also made three sets of randomer variants which we wanted to use to check the overall range of promoter strength.
For each family, we tested the randomers and the designed variants separately. To characterize the promoter strengths, we used RFP as the reporter gene and used a platereader to test for fluorescence during and after induction with IPTG.
The six designed T7 promoter variants are named as a function of their predicted promoter efficiency, relative to the wildtype. For example, T7 14 has a predicted efficiency of 14% compared to the consensus T7 promoter, whereas T7 111 is predicted to be 111% more efficient than the wildtype. In the chart above, you find each of the designed promoter variants for both the T7 with and without the lac operator, arranged in increasing predicted efficiency. Contrary to our expectation, some variants that were designed to have a lesser efficiency than the wildtype (e.g. T7 54) seem to have a much higher strength (as measured by fluorescence at saturation, normalized by the optical density). Already in the designed variants, we see a substantial difference in the behavior of the promoters that have a lac operator as opposed to those that do not. The data for this graph was produced in triplicate, so the error bar represents the standard error across those three measurements.
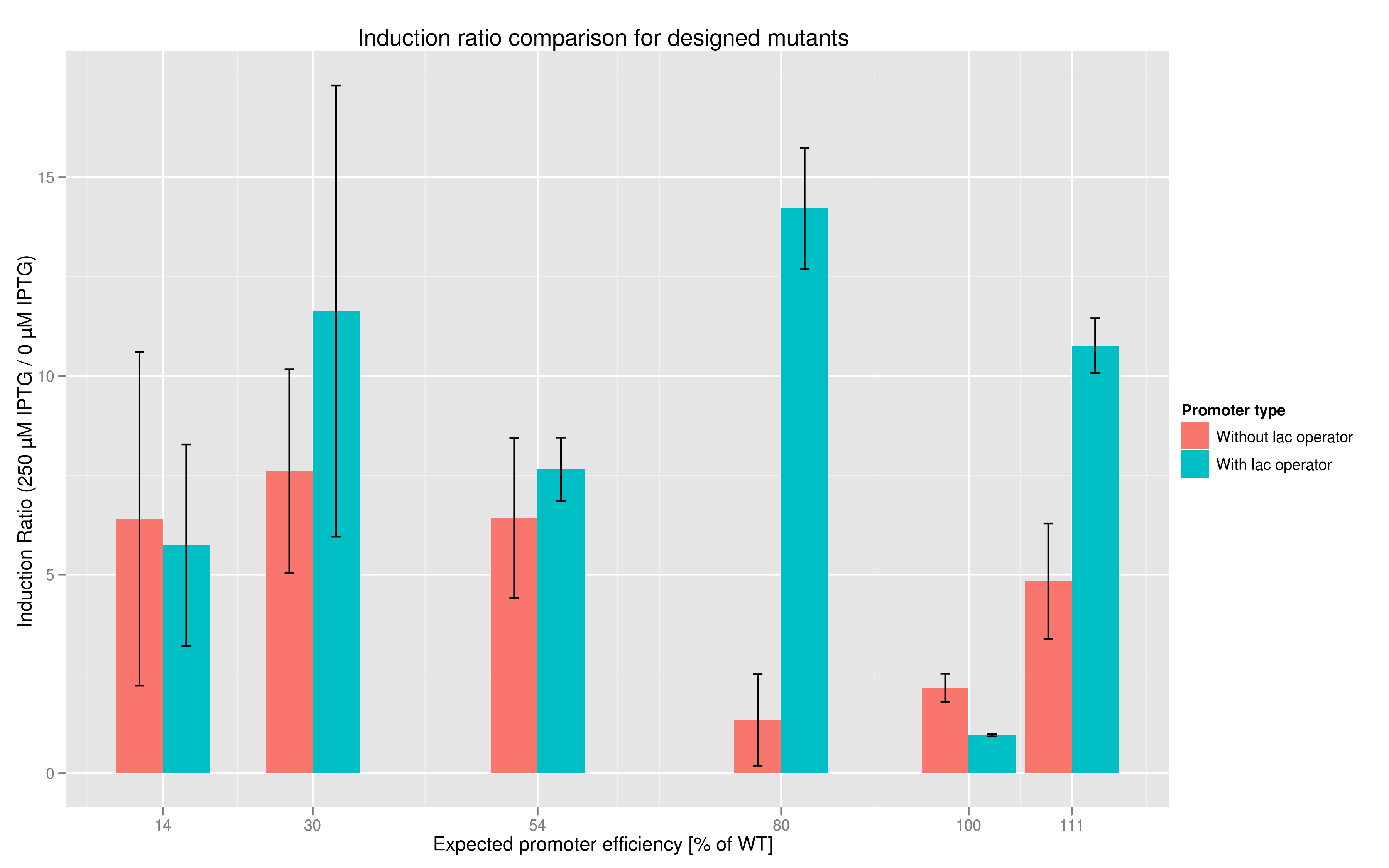
In addition to fluorescence at saturation, another way to characterize promoter strength is to look at its induction ratio, which is the ratio, at saturation, of fluorescence produced by induction with IPTG versus fluorescence produced without induction. In layman's terms, it indicates how strongly the promoter reacts to induction. Here again, our results indicate that some promoter variants (T7 80 in particular) stand out with respect to this feature. Here too the importance of the lac operator in producing high induction ratios is not to be underestimated.
For the three sets of randomers for T7 with and without the lac operator, we tested seventy-two different variants and characterized their expression using the same IPTG induction protocol as with the designed variants. The goal of using these variants was to examine the range of expressions that can be produced by random mutations as opposed to directed mutations. The results, as presented in the graph, indicate that the designed variants (with and without lac operator put together) produce a much higher average normalized fluorescence than the randomers.
Other assemblies - without new biobrick
J61002 Ptet-RFP
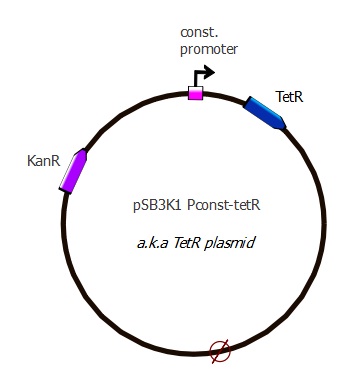
pSB3K1 Pconst-TetR
Description
This plasmid is the intermediary step before having the complete TetR plasmid. Here we only added TetR under constitutive promoter, wanting to add LacI under Ptet afterwards with a second Gibson. See the "assembly" page for more details.
Parts assembled:
- Plasmid backbone: pSB3K1 from ETHZ 2007 [http://partsregistry.org/wiki/index.php?title=Part:pSB3K1 "pSB3K1"] (taken from the delivery plate)
- Pconst: J23116 from Berkeley 2006 [http://partsregistry.org/Part:BBa_J23116 "j23116"] (sequence copied into our primers)
- RBS (B0034?) and spacer: (sequence copied into our primers)
- TetR: "TetR sequence" (623 bp) The sequence lacks a stop codon, we added TAA with our primers
Sequence
Complete sequence of the plasmid: "pSb3K1 Pconst-TetR"
Do we really need this? Sequencing data compared to the sequence of Pconst,RBS+spacer and TetR gene:"pSb3K1_TetR_seq" All these parts are correct.
Plasmid map
This plasmid contains a p15A replication origin as well as a Kanamycin resistance marker.
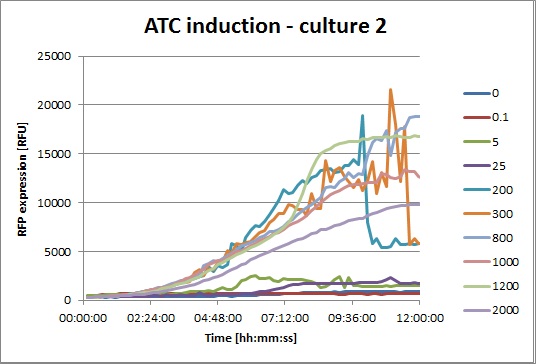
ATC induction
The platereader experiment was run for 12h, using 0-0.1-5-25-200-300-800-1000-1200-1500 nM/ul final concentrations of ATC. The cells were cotransformed with pSB3K1 Pconst-TetR and J61002 Ptet-RFP, to use the fluorescent protein as a readout for TetR inactivation by ATC. All the concentrations were tested on 4 different cultures, shown in the next graphs:
Dose-response
These data are coming from the same experiment; they show the saturation value of RFP expression for each ATC concentration. The values were averaged over the 4 different cell cultures.
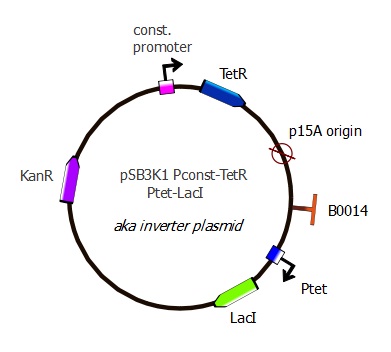
pSB3K1 Pconst-TetR Ptet-LacI
Description
This is the final TetR plasmid, containing the TetR gene as well as the LacI inverter with a Ptet promoter. Here we have wild-type TetR and Ptet sequences, but this plasmid is also intended to be used with mutant TetR genes and mutant Ptet binding sequences.
Parts assembled
- Plasmid backbone containing Pconst and TetR: see precedent section
- Terminator: B0014 from the Registry [http://partsregistry.org/Part:BBa_B0014 "B0014"] (sequence copied into our primers)
- Ptet: R0040 from Registry [http://partsregistry.org/Part:BBa_R0040 "R0040"] (sequence copied into our primers)
- LacI: amplified from Repressilator plasmid "LacI sequence" The sequence lacks a stop codon, we added TAA with our primers.
Sequence
Add results for TetR sequencing Add results for LacI asap
Plasmid map
This plasmid has the same structure as pSB3K1 Pconst-TetR: p15A replication origin and Kanamycin resistance marker. However, Ptet-LacI has been added after the p15A sequence.
ATC induction
ATC dose-response curve
IPTG induction
IPTG dose-response curve
 "
"