Team:Paris Liliane Bettencourt/Notebook/2011/09/05/
From 2011.igem.org
Danyel.lee90 (Talk | contribs) (→Digestion) |
Danyel.lee90 (Talk | contribs) (→Digestion) |
||
| Line 49: | Line 49: | ||
[[File:digestion_12_09.jpg|thumb|center|ladder 1kb - S79 - S27 - S27 - S27 - pHM3 - S105(1) - S105(2)]] | [[File:digestion_12_09.jpg|thumb|center|ladder 1kb - S79 - S27 - S27 - S27 - pHM3 - S105(1) - S105(2)]] | ||
[[File:digestion(2)_12_09.jpg|thumb|center|ladder 100 bp - S98(1) - S98(2) - S98(6) - S99(1) - S99(2)]] | [[File:digestion(2)_12_09.jpg|thumb|center|ladder 100 bp - S98(1) - S98(2) - S98(6) - S99(1) - S99(2)]] | ||
| + | |||
| + | All bands of the appropriate sizes were extracted and purified. | ||
| + | |||
| + | ===GFP amber=== | ||
| + | Quickchange mutagenesis PCR has been run. | ||
| + | |||
| + | ===Transformation=== | ||
| + | Transformation of S38 in S24. Along with the 5 ligations tubes of Babak. | ||
| + | |||
| + | ===To Do List=== | ||
| + | Check transformation results. Eventually follow-up with a PCR colony, then sequencing. | ||
| + | Digest GFP quickchange products with DpnI enzyme. | ||
| + | Check sequencing results for S92 S98 S99 | ||
| + | Read with Tecan concentration of all gel extractions. | ||
| + | Ligate & Transform : S79 into S27 and S79 into pHM3, S98 into S69, S99 into S69, S105 into S27 | ||
== Hovannes-Baptiste == | == Hovannes-Baptiste == | ||
Revision as of 21:00, 12 September 2011

Contents |
Cyrille
Miniprep
Miniprep of 2 clones of TetO array in pSB1C3, 2 clones of TetO array in pHM3 Miniprep of Mathias: S21 S82 1 2 3 4; S75 S67 and S75
With good yeilds (>500ng)
Gel Extraction
Gel extraction of pHM3 Digested in EP and pVeg YFP TetR D EP and DXP.
Ligation
Ligation of pHM3 Digested EP with pVeg-YFP-TetR D. EP with ratio 1:1 1:10 1:100 Ligation pHM3 TetO D. SP with pVeg-YFP-TetR D. SP
Transformation
Transformation of the ligation product + ComS ligated in pSB1C3 + YFP TetR BB ligated in pSB1C3
Contol of the comptent cells
Two tests of transformation was done with the B7 plasmid in TurboCell competent cells.
The rest of the tube is placed in 500µL of LB, incubated for an hour and then plated.
Danyel
Miniprep & Glycerols
- S69 (pHM3)
- S92 (Pveg_spoVG_tRNA_TT in pHM3) [Sequencing]
- S98 1,2,3,6 (pHS_tRNA) [Sequencing]
- S99 1,2,3,4 (pHS_tRNA_TT) [Sequencing]
Digestion
Everything was digested on E & P
- S69 - all 5 minipreps stored in the fridge were digested. (There seems to be a problem with the 3 pHM3 minipreps that were made today)
- S79 (pT7_RBS_GFP_TT)
- S98
- S99
- S27 (pSB1C3)
- S105 (Pveg_spoVG_T7amber_TT)
All bands of the appropriate sizes were extracted and purified.
GFP amber
Quickchange mutagenesis PCR has been run.
Transformation
Transformation of S38 in S24. Along with the 5 ligations tubes of Babak.
To Do List
Check transformation results. Eventually follow-up with a PCR colony, then sequencing. Digest GFP quickchange products with DpnI enzyme. Check sequencing results for S92 S98 S99 Read with Tecan concentration of all gel extractions. Ligate & Transform : S79 into S27 and S79 into pHM3, S98 into S69, S99 into S69, S105 into S27
Hovannes-Baptiste
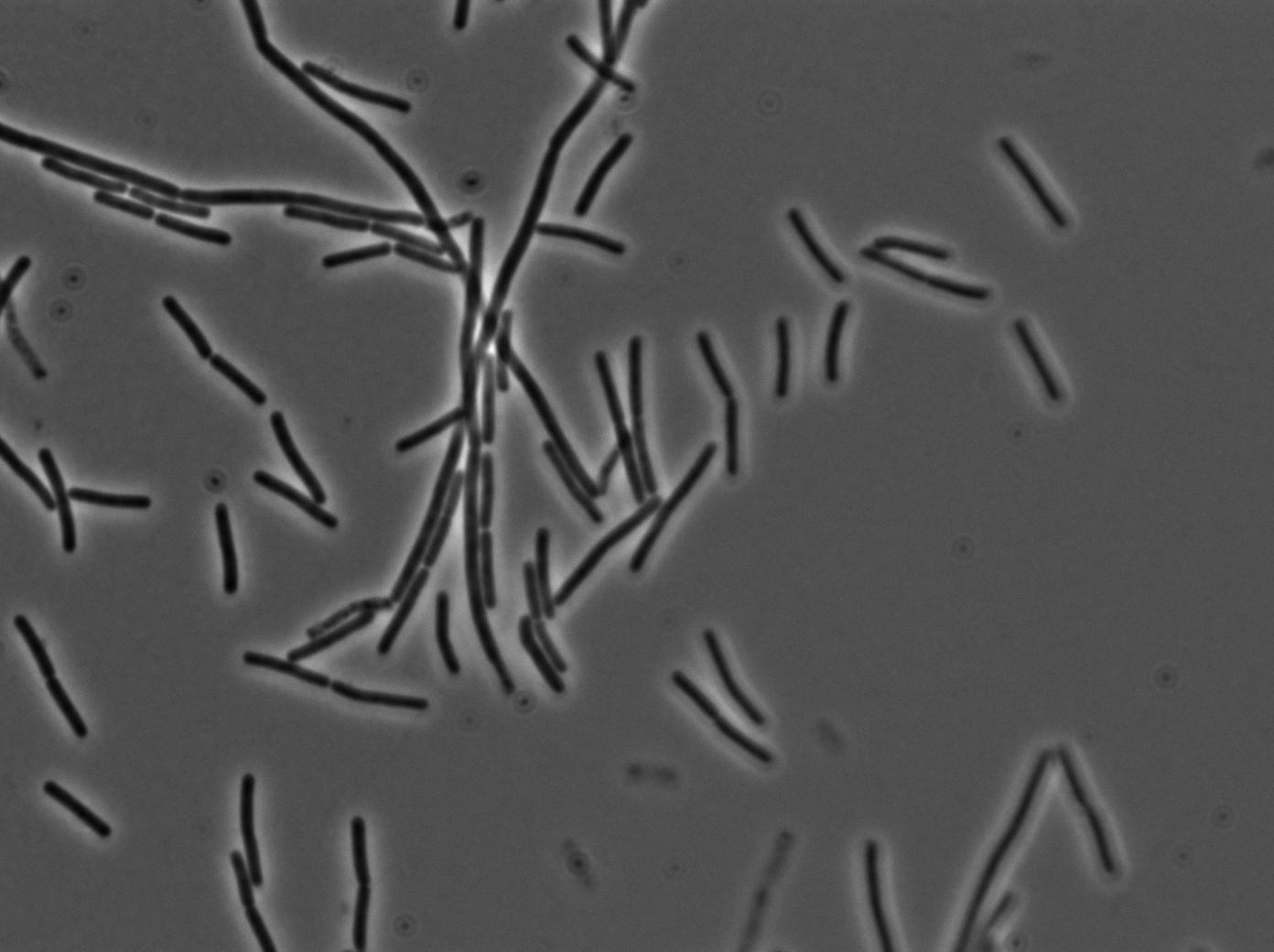
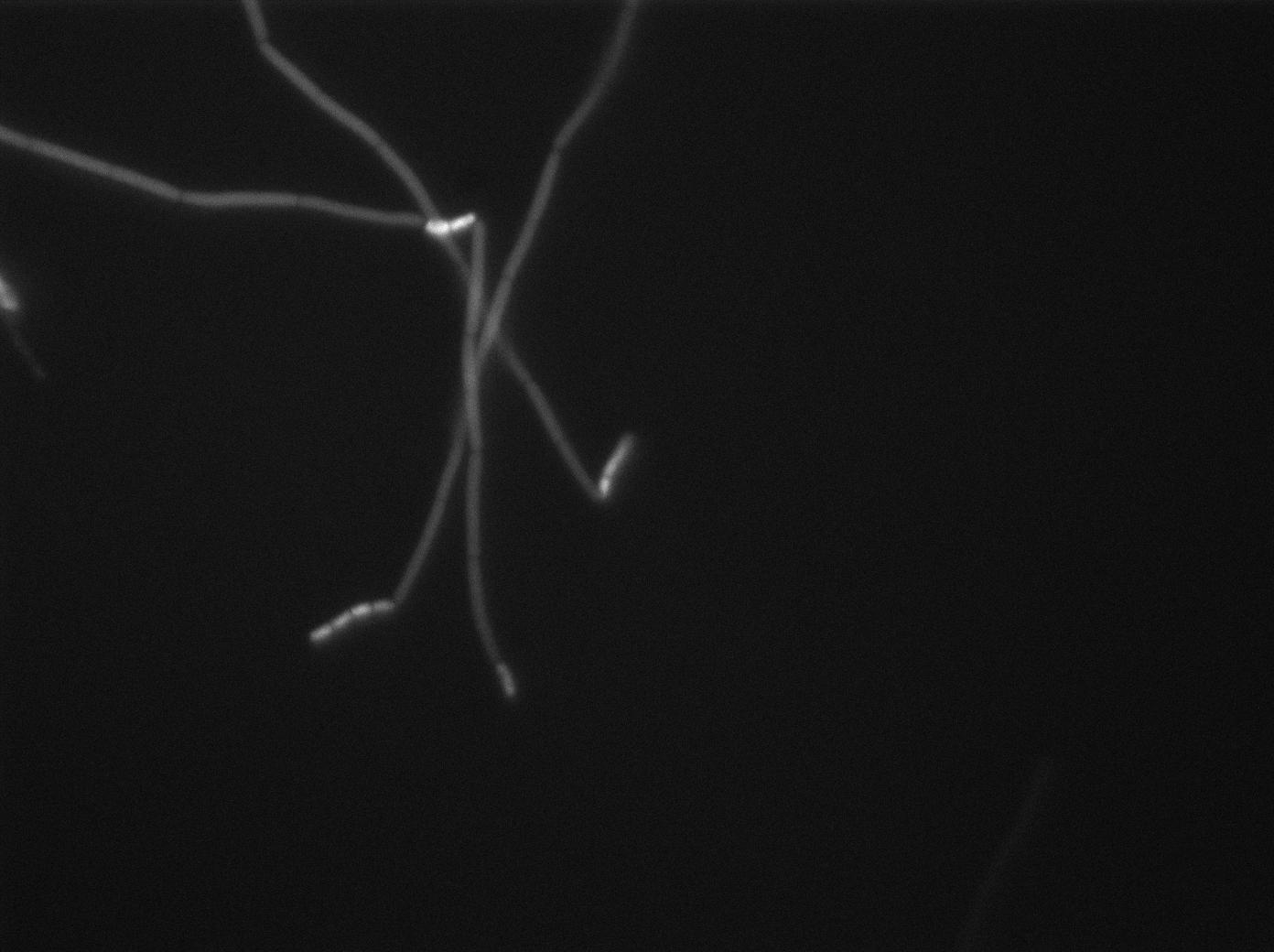
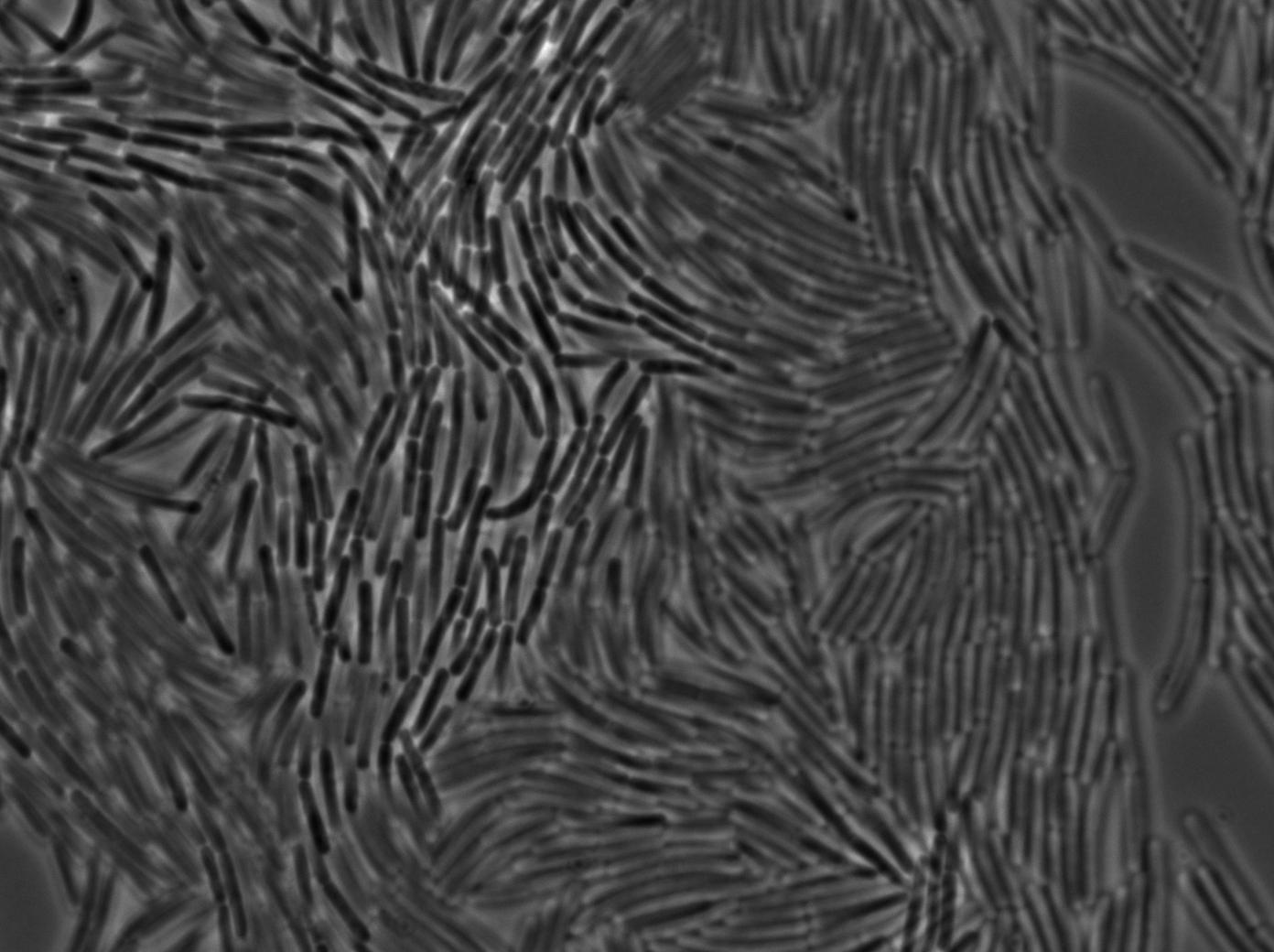
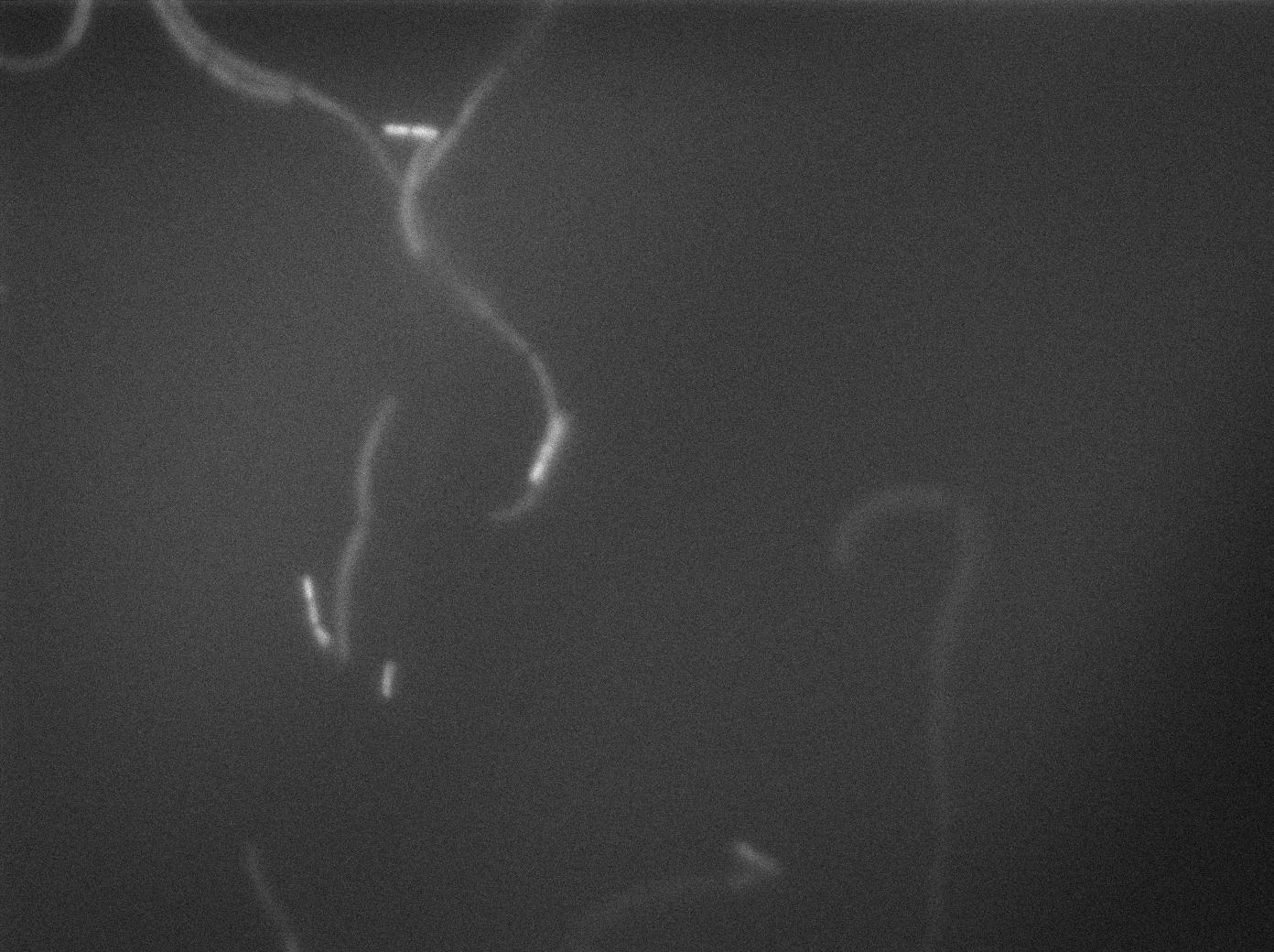
Preparation of slides
Dilution of overnight cultures : YC164 and YC227 (SinOp Design) .
YC227 has a hyperspank promoter (inducible by IPTG) which regulates a GFP gene. It also produces phosphorilated KinA.
YC164 Contains SpoA which can be indirectly phosphorilated by KinA. Phosphorilated SpoA indirectly activates a gene which contains GPF and SinI. SinI is a signal involved in biofilm shaped growth.
Two well slides : 1-control (YC164 with IPTG) 2-Mix (both strains with IPTG)
Observation
-37°C Microscopy-
We observed the plate with TRANS and YFP-filter settings on the Old Zeiss. Unfortunately, this microscope gets quickly out of focus so we need to take each picture manually. As expected, YC227 was producing GFP at the begining of the experiment, while YC164 was not. Our observations over 4 hours are the following:
- YC164 grows fast (several divisions over the experiments)
- YC227 does not grow at all and keeps its fluorescence pretty well
- YC164 does not change its state (no obvious biofilm formation)
- After letting the plate overnight we still see some florescence but not as if YC164 changed its fluorescence status.
Our conclusions
We might have poorly chosen the exact phase growth or our timing when plating the trains. Nanotube formation seems to require a more tightly packed colony. We will try this tomorrow. Biofilm formation is usually observed at lower temperatures and it could mean we will never see anything with this design (be reminded that we did not do any cloning for this construction, we only add to reuse strains from another scientific team).
 "
"