Team:EPF-Lausanne/Notebook/September2011
From 2011.igem.org
(→Tuesday, 6 September 2011) |
|||
| Line 50: | Line 50: | ||
[[File:EPFL_ATC_test_on_pSB_TetR-LacI_+_J6_Plac-RFP.jpg]] | [[File:EPFL_ATC_test_on_pSB_TetR-LacI_+_J6_Plac-RFP.jpg]] | ||
| + | Vincent transformed the "randomer" Gibson assemblies (from nearly a month ago) into BL21 cells. If you cross-check with the numerical scheme, those would be variants 7, 8, 9, 16, 17, 18. | ||
| + | |||
| + | We also went after the remaining non-randomer variants that were left (6, 13, and 14) using a colony PCR on six colonies of each variant. The PCR used 64 C for annealing and 1 minute for extension. | ||
== Wednesday, 7 September 2011 == | == Wednesday, 7 September 2011 == | ||
Revision as of 06:40, 8 September 2011
Notebook: September 2011
Contents |
Thursday, September 1 2011
Vincent PCR purified the pSB3K1-extended (no TetR) for use in the Gibsons and negative controls.
The Gibson assembly of J6-Plac-Lysis had produced a single colony. Vincent colony-PCRed it using two different sets of primers to amplify subportions of the lysis cassette. The first set of primers (30-f & 816-r) amplify an 800 bp piece whereas the second set (Seq-J6-RFP-f & Seq-J6-RFPlysis-r) amplifies a 1100 bp piece.
The gel seems to confirm that lysis is in the plasmid. We will send the plasmid for sequencing on Friday.
After taking a closer look at the plate from the platereader, it seems that none of the T7 variants did any lysing. This realization made it urgent that we discover whether or not the full T4-lysis cassette was really in these Gibson-assembled plasmids. The samples that had been sent in for sequencing had primers that seemed to amplify the backbone, instead of the lysis cassette. While this does not necessarily mean that the lysis is not there, it does suggest that all the PCRS and gels used to verify the presence of lysis were only showing the existence of the backbone.
Alina ran a PCR with different primers from Douglas' first attempts at putting together the 2700 bp cassette that amplify select regions of the cassette.
Friday, September 2 2011
Alina wanted to try a new extension PCR of the T4-lysis and a Gibson of the exteneded-lysis into the K1 backbone. The transformation was successful with tons of colonies. A colony PCR will be run on Monday.
Nadine came back in the lab and cotransformed pSB3K1 Pconst-TetR with J61002 Plac-RFP since the sequencing results were concluent, in order to perform IPTG and ATC platereader experiments.
We had a lot of contaminated bottles, so she also made new SOC bottles.
Monday, September 5 2011
Alina ran a colony PCR on the K1-T7-lysis Gibson assembly.
After having set liquid cultures on Saturday, Nadine set up the platereader experiment as follows:
- J61002 Plac-RFP with increasing concentrations of IPTG
- pSB3K1 TetR-LacI + J61002 Plac-RFP with increasing concentrations of IPTG
- pSB3K1 TetR-LacI + J61002 Plac-RFP with increasing concentrations of ATC
She had also prepared co-transformations with pSB3K1 TetR-LacI + J61002 Plac-lysis, but the sequencing results indicated that the lysis cassette was wrong.
Tuesday, 6 September 2011
The results from the platereader experiment came back and they look good!
- J6 Plac-RFP: The level of RFP expression doesn't rise much with high doses of IPTG, showing that DH5alpha cells don't express much LacI and that Plac is a quite weak promoter.
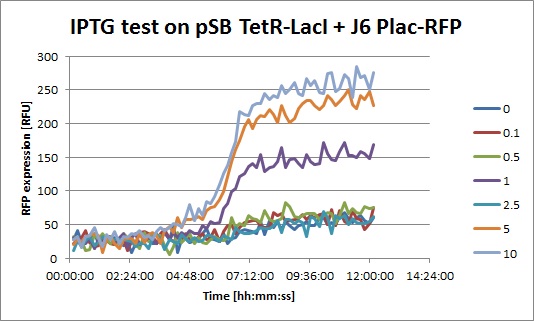
- J6 Plac-RFP + pSB TetR-LacI, IPTG: you can clearly see an increase of RFP when IPTG is added. This means that even with a constitutive promoter in front of TetR, LacI is still expressed. In fact, Pconst is weaker than Ptet. The "2.5" data seems strange, I perhaps missed to put IPTG in this well.
- J6 Plac-RFP + pSB TetR-LacI, ATC: Without ATC, we have a similar expression curve as in the IPTG experiment. When ATC is added, we see a diminution of RFP expression because LacI is not repressed anymore.
Vincent transformed the "randomer" Gibson assemblies (from nearly a month ago) into BL21 cells. If you cross-check with the numerical scheme, those would be variants 7, 8, 9, 16, 17, 18.
We also went after the remaining non-randomer variants that were left (6, 13, and 14) using a colony PCR on six colonies of each variant. The PCR used 64 C for annealing and 1 minute for extension.
Wednesday, 7 September 2011
Nadine made colony PCRs for the T7 promoter variants that Henrike cloned into the biobrick vector. She also prepared some liquid cultures for tomorrow's platereader experiment.
 "
"