Team:Bielefeld-Germany/Labjournal
From 2011.igem.org
(Difference between revisions)
(→Week 3: 16th - 22nd may) |
|||
| Line 6: | Line 6: | ||
* cloning of fusionprotein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter | * cloning of fusionprotein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter | ||
* testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection | * testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection | ||
| - | * expression of the successfully assembled BPA degrading BioBricks in ''E. coli'' TOP10 | + | * expression of the successfully assembled BPA degrading BioBricks in ''E. coli'' [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] |
S-layer: | S-layer: | ||
| Line 23: | Line 23: | ||
S-layer: | S-layer: | ||
| - | * expression of the S-layer gene without TAT-sequence of ''B. flavum'' in ''E. coli'' KRX and BL21-Gold(DE3) after sequencing gave correct results | + | * expression of the S-layer gene without TAT-sequence of ''B. flavum'' in ''E. coli'' KRX and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results |
* successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 ''Corynebacterium halotolerans''] | * successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 ''Corynebacterium halotolerans''] | ||
Revision as of 17:38, 20 May 2011
On this page we summarize the (successful) results and achievements of our teamwork.
Week 1: 2nd - 8th may
Bisphenol A:
- cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic)
- cloning of fusionprotein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter
- testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection
- expression of the successfully assembled BPA degrading BioBricks in E. coli [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10]
S-layer:
- successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html Brevibacterium flavum] and Corynebacterium crenatum
- successful cloning of the S-layer gene of B. flavum without TAT-sequence
Organizational:
- meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld
Week 2: 9th - 15th may
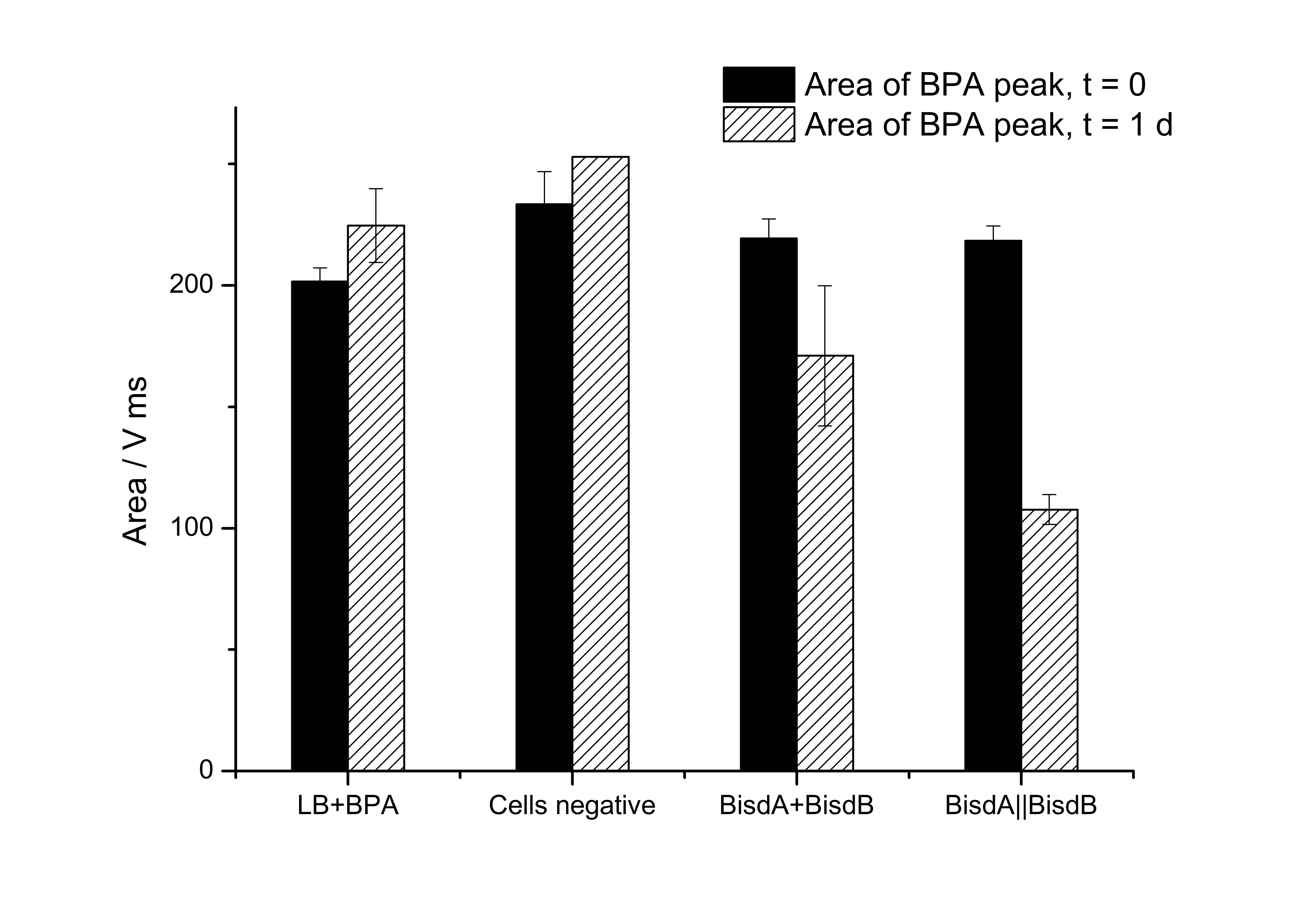
Figure 1: HPLC results of the first experiment on BPA degradation in E. coli TOP10. Cultivations were carried out in LB medium with 100 mg L-1 BPA at 30 °C. Samples were taken at the beginning of the cultivation and after one day. The HPLC results are shown above (area of the BPA peak). BisdA + BisdB is the polycistronic gene and BisdABisdB is the fusion protein.
Bisphenol A:
- assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system)
- HPLC results: fusionprotein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare fig. 1)
S-layer:
- expression of the S-layer gene without TAT-sequence of B. flavum in E. coli KRX and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results
- successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 Corynebacterium halotolerans]
Organizational:
- moving to our own room in the [http://www.cebitec.uni-bielefeld.de/ CeBiTec]
Week 3: 16th - 22nd may
Bisphenol A:
- successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP oxidoreductase] gene from E. coli TOP10
S-layer:
- successful cloning of the complete S-layer genes of B. flavum, C. crenatum and C. halotolerans
- successful cloning of the S-layer gene of B. flavum without TAT-sequence, without lipid anchor and without both (only self-assembly domain)
- successful cloning of the S-layer gene of C. halotolerans without TAT-sequence and lipid anchor (only self-assembly domain)
Organizational:
 "
"