Team:EPF-Lausanne/Notebook/August2011
From 2011.igem.org
(→Thursday, 04 August 2011) |
|||
| Line 51: | Line 51: | ||
== Thursday, 04 August 2011 == | == Thursday, 04 August 2011 == | ||
| - | The mutant TetR plasmids plated yesterday yielded only one colony, even the mutation control didn't work. | + | The mutant TetR plasmids plated yesterday yielded only one colony, even the mutation control didn't work. Nadine plated the rest of transformed cells, but probably the cells in the mutagenesis are too old to be competent. |
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Revision as of 14:01, 4 August 2011
Notebook: August 2011
Tuesday, 02 August 2011
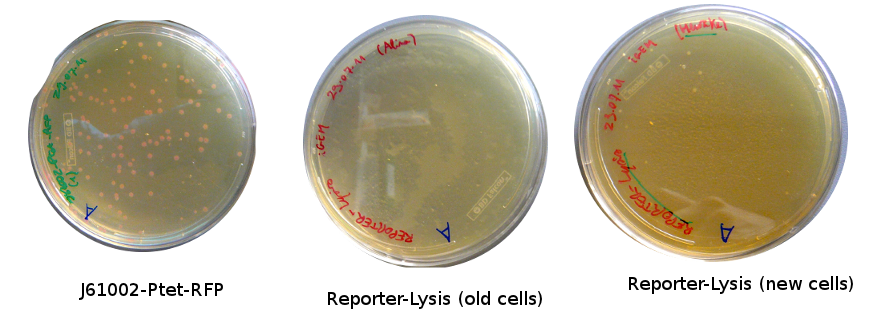
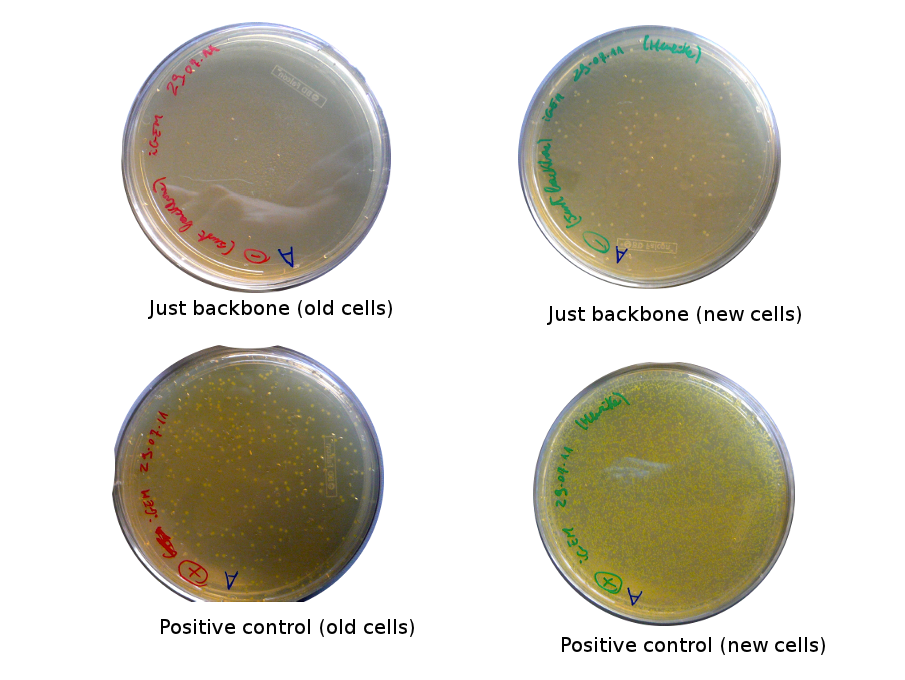
Lilia came on Saturday to take the plates and here are the results:
The old competents cells seem not to work very well and the Gibson assembly failed since in the plate with only backbone we have a lot of cells therefore the backbone can closes on itself.
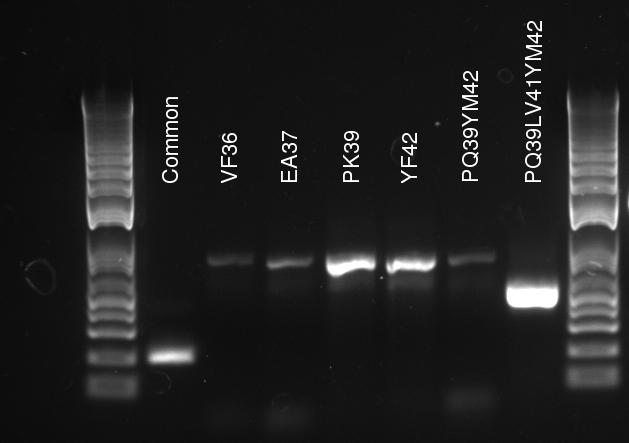
Alessandro make the PCR (gradient PCR in which the annealing temperature increases for each sample) to amplify TetR (putting Pconst in front of it) from Repressilator plasmid to make the Gibson for the Pconst-TetR plasmid. The result is illustrated on the right.
Since we have already amplified the backbone (pSB3K1) we can make the Gibson assembly to make the Pconst-TetR plasmid.
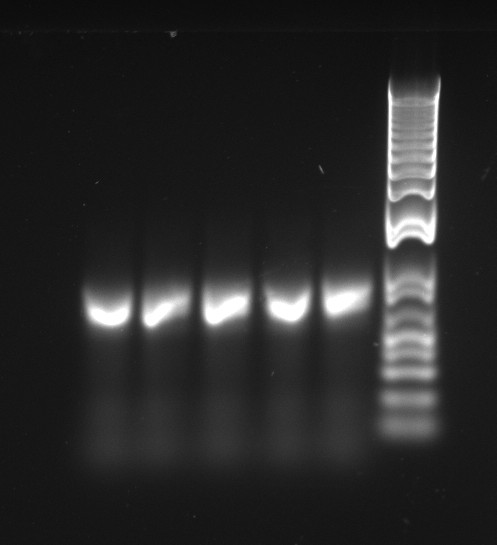
The latest mutation PCR yielded large amounts of product. The two bands might be a little worrying, but overall this is a lot better than any of the previous trials, with the old polymerase. The stitch PCR will be ran tomorrow.
On the side, more reporter and lysis plasmids were assembled (together with a control, containing only the backbone), from the previously made gibson fragments (PCR purified on 2nd August 2011).
Wednesday, 03 August 2011
Alessandro made glicerol stocks of Vincent's plasmid (J61002-Ptet-RFP) and purified the PCR product of the day before (Pconst-TetR) to make the Gibson assembly (using the already amplified backbone of pSB3K1). The Gibson assembly for the pSB3K1-Pconst-TetR (using either equimolar ratio of backbone-insert either 2 times insert) and for Reporter-RFP/Lysis has been plated.
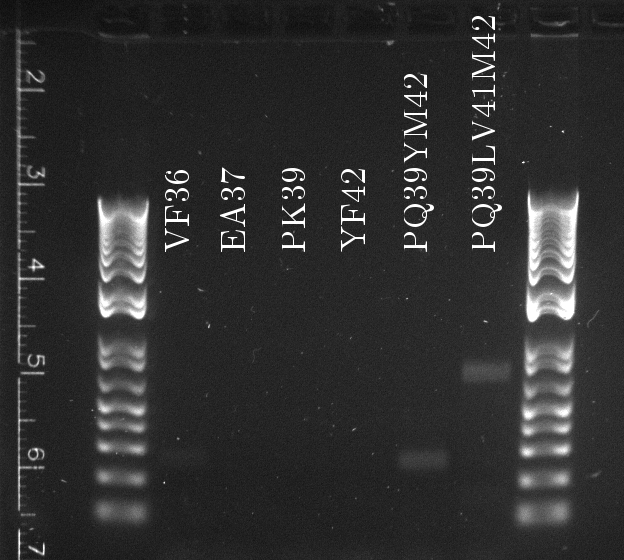
Douglas ran the stitch PCR on the latest mutants, unfortunately yielding virtually no product. Only the PQ39LV31YM42 mutant has detectable amounts of product in the correct size, but then it also had a much larger concentration of template (or so it seems by eyeballing the mutation PCR's gel).
Likely causes:
- The common sequence DNA was lost during PCR purification (Doug accidentally used the high cut-off buffer), in which case the mutation PCR should be repeated from scratch...at least for the common sequence.
- The 3' final and 5' final primers are not suitable for the stitch step. Concentrations and heat cycles should be accurate, because they worked for the mutation PCR.
Nadine transformed and plated Doug's mutant TetR plasmids from 26.07. She prepared the X-gal and IPTG solutions needed for the control, but they still need to be filtered.
We tried a first colony PCR on J61002 Ptet-RFP cells, but nothing got amplified... Tomorrow we'll try the primers on purified plasmids to see it the problem comes from here.
Thursday, 04 August 2011
The mutant TetR plasmids plated yesterday yielded only one colony, even the mutation control didn't work. Nadine plated the rest of transformed cells, but probably the cells in the mutagenesis are too old to be competent.
 "
"