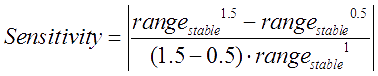Team:Peking S/modeling/Parameters
From 2011.igem.org
| Line 37: | Line 37: | ||
<html> | <html> | ||
</div> | </div> | ||
| - | Top↑< a href=“#top” | + | Top↑< a href=“#top”> |
</html> | </html> | ||
Revision as of 08:24, 5 October 2011
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Population Density Balancer
Parameters
There are over 30 parameters in our model, which increases the difficulty of modeling conduction. This is similar to what we encountered in 2009 iGEM competition. Therefore we adopt some assumptions from 2009 Peking iGEM modeling work. According to those assumptions, we can decide either the value or the range of the parameters. Nevertheless, parameter c1 and c2 were not included, and we do not know their estimated value. Therefore, we sampled these two parameters in a quite wide range ( ) to guarantee that the appropriate value lies within sampling range.
To verify the credibility of the parameters values, we analyzed the sensitivity of parameters by following definition (the variation range of A cell population in steady state was employed to indicate the performance of the balancer): Each parameter varied respectively while other parameters fixed, then the extent of system performance variation was observed to calculate the sensitivity of each parameter.
The smaller the sensitivity value is, the more reasonable the parameters are.
Here we provide a graph to show the effect of parameter variation on the cell population dynamics.
From the result of sensitivity analysis, we found that the sensitivity of our parameters is relatively low, while some of them are still sensitive enough, capable of tuning the proportion of A/B cell in the total population.
 "
"