Yeast Vector Library
7/1/11
- Made growth plates for the 12 vectors:
- pRS 403: HIS3 Selectable, Integrating
- pRS 404: TRP1 Selectable, Integrating
- pRS 405: LEU2 Selectable, Integrating
- pRS 406: URA3 Selectable, Integrating
- pRS 413: HIS3 Selectable, CEN/ARS Episomal
- pRS 414: TRP1 Selectable, CEN/ARS Episomal
- pRS 415: LEU2 Selectable, CEN/ARS Episomal
- pRS 416: URA3 Selectable, CEN/ARS Episomal
- pRS 423: HIS3 Selectable, 2micron Episomal
- pRS 424: TRP1 Selectable, 2micron Episomal
- pRS 425: LEU2 Selectable, 2micron Episomal
- pRS 426: URA3 Selectable, 2micron Episomal
- Repipetted primers
- Started PCR experiment design
7/2/11
- Created 2 mL overnight cultures of vectors in E. coli w/ LB + Carbenicillin
- 37C incubation
7/3/11
Miniprep 12 vectors from 2 mL overnights:
- Use Zymo's Zyppy Kit Mini prep kit.
- Store in -20C
7/5/11
- Designed the multiple cloning site substitution and QuickChange protocol. For site-directed mutagenesis, primers must be completely complementary to each other with the base change in the exact middle of the primer. 15-20 bp of homology on either side of the base change is ideal. For a large bp substitution (in this case, the replacement of a multiple cloning site), the primers need regions of homology on both the 5' and 3' strands (5' for primer 1, 3' for primer 2) on either side of the site of the change, with the desired sequence change complementary in both primers.
7/6/11
- Created and consolidated all PCR reagents and DNA for tomorrow's PCR
7/7/11
- Multiple cloning site substitution PCR
7/8/11
DpnI Digest:
| PCR Product | 50 µL |
| DpnI | 1 µL |
| 10x NEBuffer 2 | 5 µL |
| Total | 50 |
- Digest overnight
7/11/11
Transformation:
| Competent cells | 50 µL |
| Digested PCR product | 5 µL |
- Thaw cells on ice
- Gently pipet PCR product into competent cells
- 30 min incubation on ice
- Heat shock 45 sec @ 42C
- Add 500µL SOC and incubate at 37C for hr
- Plate 450µL and save 50µL
7/12/11
- No colonies
- Repeat transformation and plating
7/13/11
- No colonies
- Optimized PCR protocol according to Agilent's Herculase manual
7/14/11
- Regrew original pRS vectors in overnight cultures in case minipreps are bad.
7/15/11
- Mini prep vectors
- MCS delete/insert PCR
7/16/11
- PCR Purification
7/18/11
- DpnI digest
- Overnight ligation at 16C
7/19/11
- Transformation of 12 vectors
7/20/11
- No colonies
- Verify original vectors with EcoRI digest
- Verification successful
7/21/11
- Re-do transformation
7/22/11
- Transformation failed
- Change QCPCR protocol to new two-step process
- Create competent cells
7/25/11
- Try new Site Directed Mutagenesis QCPCR protocol on pRS 416 @ various template and primer concentrations
7/26/11
- Yeast spec. assay
- DpnI digest
- 416 Site Directed Mutagenesis Transformation
7/27/11
- Bad competent cells (no colonies on pos. ctrl.)
- Redo QCPCR w/ 9 primer/template concentrations
7/28/11
- DpnI digest of 416 QCPCR
- Transformation using high efficiency TOP10 cells
7/29/11
- 416 SDM QCPCR worked!
- Grow overnight cultures
8/1/11
- Miniprep
- 2nd step QCPCR
8/2/11
- DpnI digest
- Transformation
8/3/11
- Transformation successful: finished 416 biobricked vector
- Grow overnight culture
8/4/11
- Create new competent cells using TSS protocol
8/25/11
- 1st QCPCR on rest of vectors
- DpnI digest
- Transformation
8/26/11
- Colonies for all but 413 and 403
- Grow overnight cultures
8/27/11
- Miniprep successful transformations
8/29/11
- 2nd QCPCR on successful 1st QCPCRs
8/30/11
- DpnI digest
8/31/11
- QCPCR (site directed mutagenesis) on 403, 405, 413, 415, 423
9/1/11
- Digest EcoRI, XbaI, SpeI and PstI on final biobricked vector (416)
- Run Gel: Failed (PstI had 3 products instead of 1)
9/5/11
- DpnI digest 8/31 reactions
- Transform 8/31 reactions
9/6/11
- Colonies for 403, 405, 413, 415, 423, 424, 425
9/7/11
- Redo first second round QCPCR (multiple cloning site swap) 9/6 colonies
9/8/11
- PCR failed: Samples evaporated in PCR machine
9/12/11
- Redo first round (site directed mutagenesis) on all pRS vectors
9/13/11
- DpnI digest
- Transform 9/7 QCPCR
9/14/11
- No colonies
- Problem may be in incompatible PCR buffer (Herculase buffer) and Transformation buffer. Solution is to do PCR purification before transformation.
- Check theory by running PCR purification on PCR products that previously failed transformation and re-transform
9/15/11
- Colonies! Theory confirmed.
- Re-run all QCPCR (site directed mutagenesis) reactions
9/16/11
- DpnI digest
- Transform
9/17/11
- All QCPCR reactions had large amount of colonies
- Miniprep all samples
9/20/11
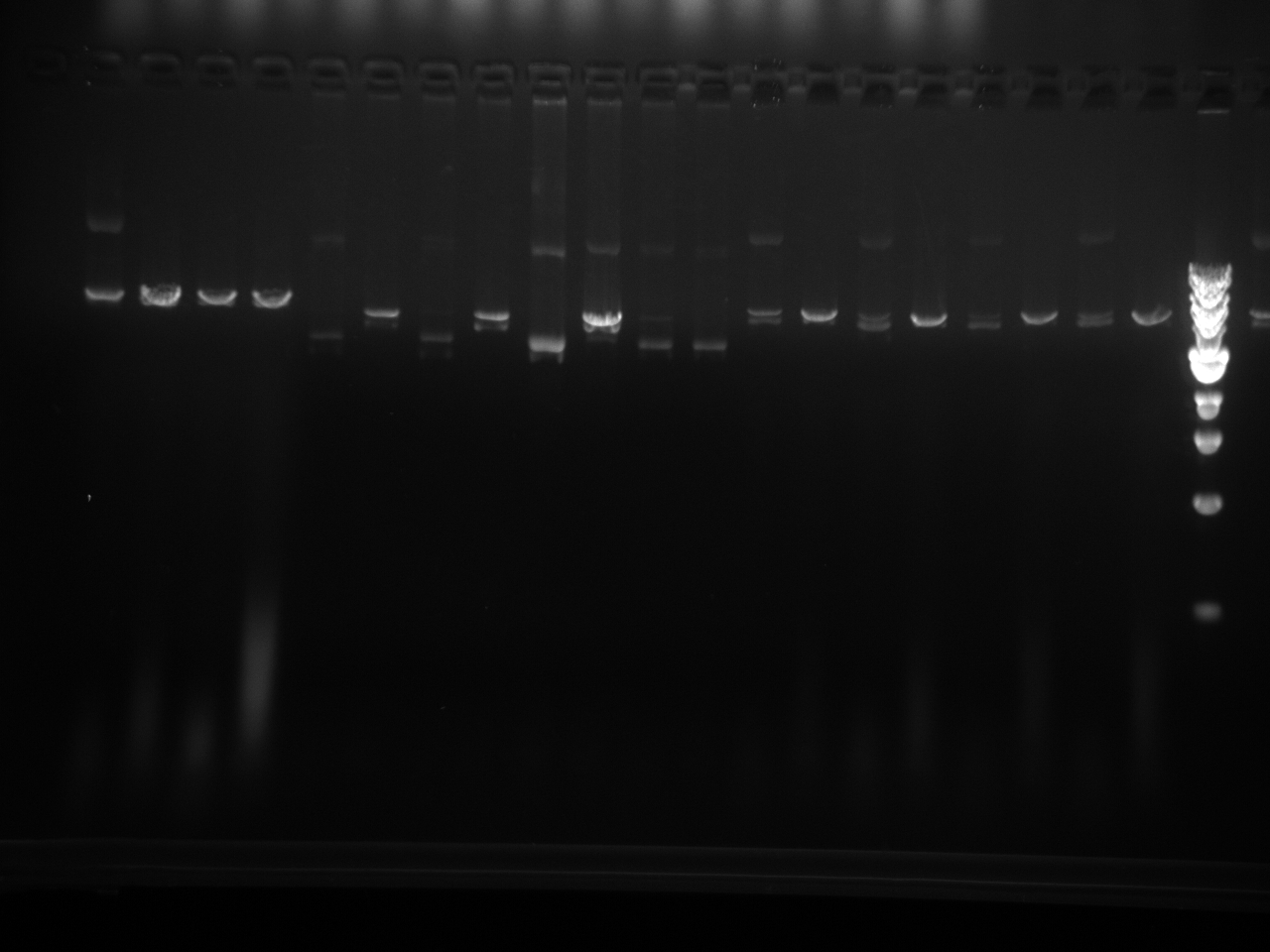
Restriction enzyme digest check on all miniprepped vectors:
| DNA | 2 µL |
| Restriction Enzyme | 0.5 µL |
| 10x NEBuffer 2 | 1 µL |
| DI Water | 6.5 µL |
| Total | 10 µL |
Restriction Digest results: We are looking for 1 band here: linearization of the vector. The - sign after the vector names means it has completed site directed mutagenesis QCPCR. It can go on to the final QCPCR, multiple cloning site swap.
9/21/11
- The rest of the site-directed mutagensis QCPCRs need to be done using the optimized 2-stage approach.
- DpnI overnight digest
9/22/11
- PCR purification
- Transformation on LB/Carb plates using previously mentioned protocol
9/23/11
- Pick colonies
- Overnight cultures
9/24/11
- Miniprep
- Gel verification
9/25/11
- QCPCR final (MCS substitution) on final vectors
- DpnI overnight digest
9/26/11
- PCR purification
- Transformation
9/25/11
- Pick colonies
- Overnight cultures
9/26/11
- Miniprep: Yield too low
9/27/11
- Re-miniprep
9/28/11
- Gel verification
Looking for alternating uncut/cut bands. This is because we cut with a restriction enzyme in the old multiple cloning site before we substituted in the BioBrick MCS. The other band is a restriction enzyme site present in the final version of the vector.
 "
"