Team:Arizona State
From 2011.igem.org
| Line 2: | Line 2: | ||
We are Arizona State University's first iGEM team, working over the summer for the 2011 International Genetically Engineered Machine competition. | We are Arizona State University's first iGEM team, working over the summer for the 2011 International Genetically Engineered Machine competition. | ||
| - | + | [[Image:ASU_tips.png|400px]] | |
== Abstract == | == Abstract == | ||
<p>Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are a genomic feature of many prokaryotic and archaeal species. CRISPR functions as an adaptive immune system, targeting exogenous sequences that match spacers integrated into the genome. Our project focuses on developing a set of tools for synthetic control over the CRISPR pathway. This includes a method for creating polymers of repeat-spacer-repeat units, the development of CRISPR biobricks (CAS genes, leader sequences) for several CRISPR subtypes (E. coli, B. halodurans, and L. innocua), testing these components on plasmids containing GFP, and a software tool to collect and display CRISPR information, as well as select spacers from a particular sequence. Given the relatively recent progress in the scientific understanding of this system, we see the potential for a wide range of biotechnological applications of CRISPR in the future.</p> | <p>Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are a genomic feature of many prokaryotic and archaeal species. CRISPR functions as an adaptive immune system, targeting exogenous sequences that match spacers integrated into the genome. Our project focuses on developing a set of tools for synthetic control over the CRISPR pathway. This includes a method for creating polymers of repeat-spacer-repeat units, the development of CRISPR biobricks (CAS genes, leader sequences) for several CRISPR subtypes (E. coli, B. halodurans, and L. innocua), testing these components on plasmids containing GFP, and a software tool to collect and display CRISPR information, as well as select spacers from a particular sequence. Given the relatively recent progress in the scientific understanding of this system, we see the potential for a wide range of biotechnological applications of CRISPR in the future.</p> | ||
Revision as of 02:52, 29 September 2011
|
|
We are Arizona State University's first iGEM team, working over the summer for the 2011 International Genetically Engineered Machine competition.
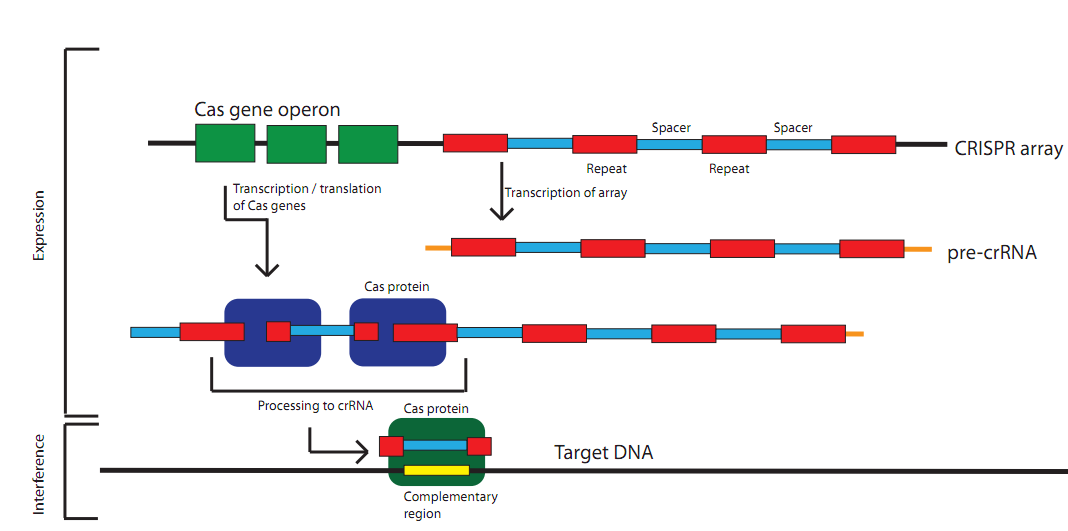
AbstractClustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are a genomic feature of many prokaryotic and archaeal species. CRISPR functions as an adaptive immune system, targeting exogenous sequences that match spacers integrated into the genome. Our project focuses on developing a set of tools for synthetic control over the CRISPR pathway. This includes a method for creating polymers of repeat-spacer-repeat units, the development of CRISPR biobricks (CAS genes, leader sequences) for several CRISPR subtypes (E. coli, B. halodurans, and L. innocua), testing these components on plasmids containing GFP, and a software tool to collect and display CRISPR information, as well as select spacers from a particular sequence. Given the relatively recent progress in the scientific understanding of this system, we see the potential for a wide range of biotechnological applications of CRISPR in the future. What is CRISPR?Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are a genomic feature of many prokaryotic and archeal species. CRISPR functions as an adaptive immune system. A CRISPR locus consists of a set of CAS (CRISPR associated) genes, a leader, or promoter, sequence, and an array. This array consists of repeating elements along with "spacers". These spacer regions direct the CRISPR machinery to degrade or otherwise inactivate a complementary sequence in the cell.
|
 "
"