Team:Arizona State/Project/Software
From 2011.igem.org
Rubenacuna (Talk | contribs) (Added images.) |
Rubenacuna (Talk | contribs) (Added information about the source.) |
||
| Line 23: | Line 23: | ||
'''Source''' | '''Source''' | ||
---- | ---- | ||
| - | * | + | * The source code is available on our [http://code.google.com/p/crispr-studio/ GoogleCode] page. It is available as a package of the latest revision and directly from Subversion. |
| - | :* Dependencies: BioPython, NumPy, WxPython, BLAST+ | + | :* Dependencies: [http://biopython.org/wiki/Download BioPython] 2.7.2 or compatible, [http://new.scipy.org/download.html NumPy] 1.6.1 or compatible, [http://www.wxpython.org/download.php WxPython] 2.8 or compatible, [ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ BLAST+] 2.2.25 or compatible |
| + | |||
| + | <p>Pick os/arch as needed. Note that each of the python packages should list the version of Python they support - select the right one. NumPy must be installed before BioPython. The x64 release of wxPython doesn't seem to like Win7 so it may be better to use the x86 release. BLAST+ must be included in the system path. If BLAST+ is not available in the the system, the spacer creator will not function. The information browser will still function normally.</p> | ||
| + | |||
| + | <p>The code itself is not fully documented. However, the code was intentionally written to be readable without comments. Project files are available for PyCharm 1.6.</p> | ||
| + | |||
| + | <p>You may then run MainFrame.py to start the program.</p> | ||
'''Interface''' | '''Interface''' | ||
Revision as of 00:24, 28 September 2011
|
|
We have developed a tool to assist in the development of synthetic CRISPR systems.
Downloads
Installation Using the provided binaries is the fastest way to get started with CRISPRstudio. Visit our GoogleCode page and select the downloads tab. Download the appropriate build based on your OS. All builds are provided for x87 platforms. For x64 real support, you will need to run the source directly. x64 support has not shown significant speed increases for CRISPRstudio. Extract the compressed file to some location. You also need to download BLAST+ 2.2.25 or compatible. BLAST+ must be installed and included in the system path. On most systems, the BLAST+ installer will take care of this. After this, you may run MainFrame to start the program. Source
Pick os/arch as needed. Note that each of the python packages should list the version of Python they support - select the right one. NumPy must be installed before BioPython. The x64 release of wxPython doesn't seem to like Win7 so it may be better to use the x86 release. BLAST+ must be included in the system path. If BLAST+ is not available in the the system, the spacer creator will not function. The information browser will still function normally. The code itself is not fully documented. However, the code was intentionally written to be readable without comments. Project files are available for PyCharm 1.6. You may then run MainFrame.py to start the program. Interface 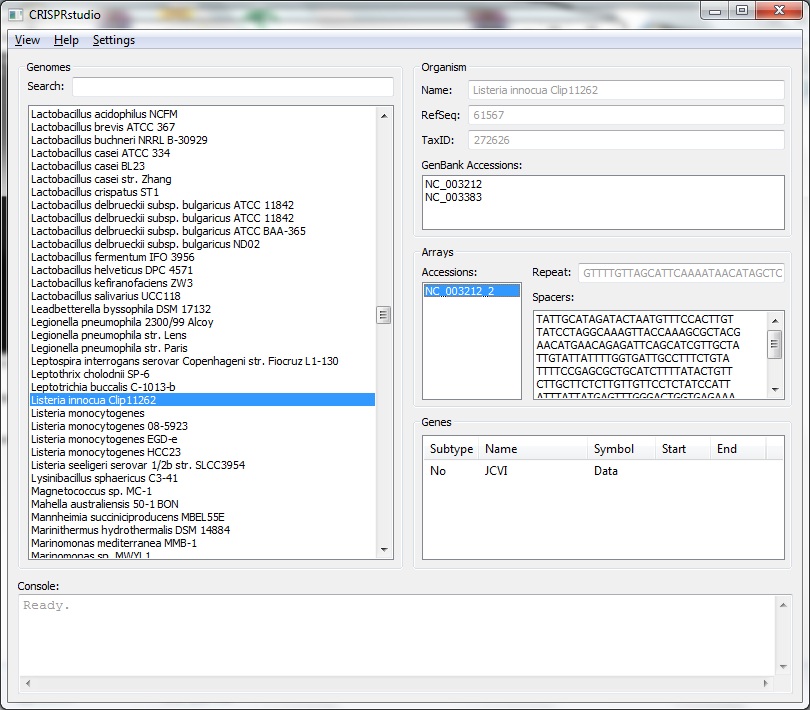 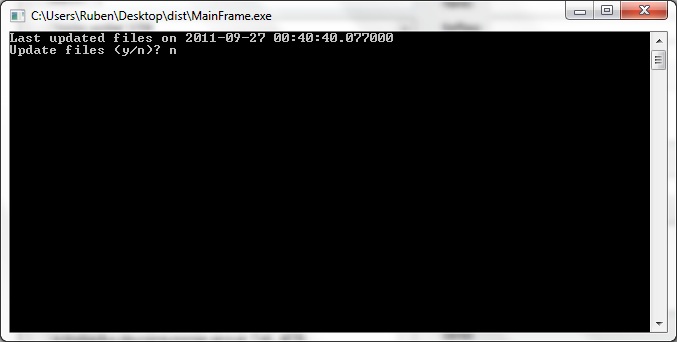  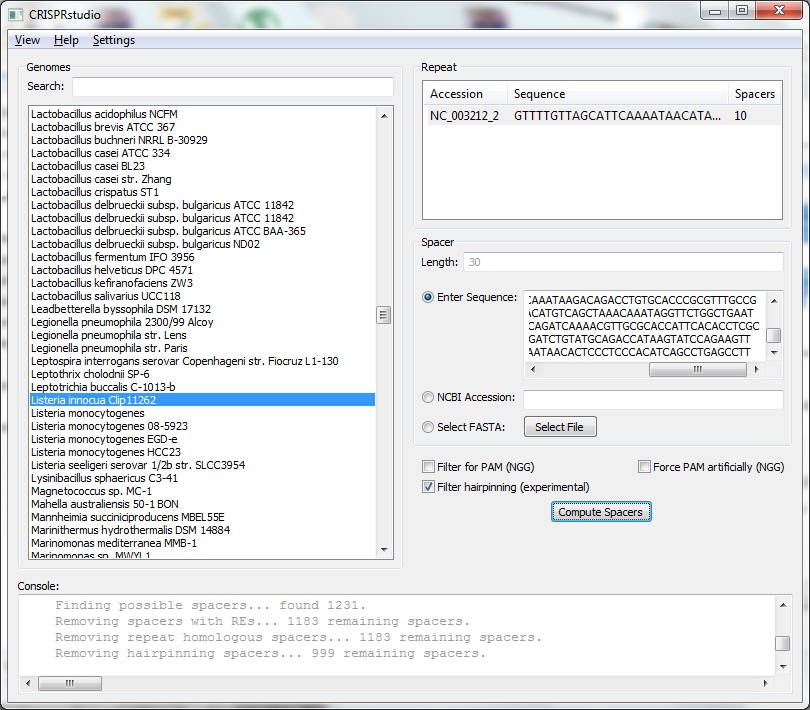 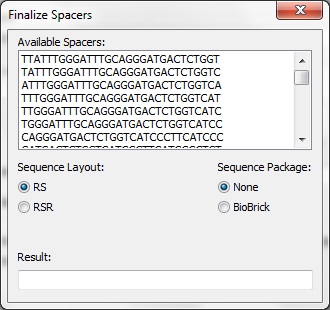 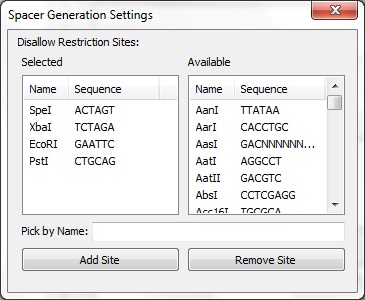
|
 "
"
