Team:UCL London/Research/Stresslights/Experiments
From 2011.igem.org
| Line 7: | Line 7: | ||
<div id="content"> | <div id="content"> | ||
<h1>Modifying Oximeter in the lab</h1> | <h1>Modifying Oximeter in the lab</h1> | ||
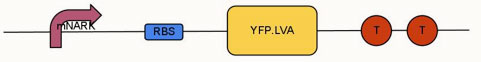
| - | + | '''Aim:''' To modify and improve the oximeter device (BBa_K239011) using modified nark promoter (BBa_K239006) submitted by 2009 UCL iGEM team. Changes included replacing the normal GFP (green fluorescent protein) with an enhanced YFP (yellow fluorescent protein), which has an LVA tag added to its C terminal. | |
| - | To modify and improve the oximeter device (BBa_K239011) using modified nark promoter (BBa_K239006) submitted by 2009 UCL iGEM team. Changes included replacing the normal GFP (green fluorescent protein) with an enhanced YFP (yellow fluorescent protein), which has an LVA tag added to its C terminal. | + | |
[[File:Ucl-content-Oximieter-circuit.jpg|center]] | [[File:Ucl-content-Oximieter-circuit.jpg|center]] | ||
| Line 106: | Line 105: | ||
</tr> | </tr> | ||
</table></div></html> | </table></div></html> | ||
| + | |||
| + | <h2>Characterisation Plan for the new improved Oximeter</h2> | ||
| + | '''Aim:''' To compare the sensitivity and half-life of fluorescence assays between Oximeter 2.0 (mNark/RBS/YFP.LVA/Terminator on pSB1C3) and Oximeter 1.0 (mNark/RBS/YFP/Terminator on pSB1C3) | ||
| + | |||
| + | # Carry out 3A assemblies to construct the Oximeter 1.0 device with wild type YFP (yellow fluorescent protein) | ||
| + | # Transform two separate stocks of competent TOP10 E. coli cells with the mini-prep DNA for Oximeter 1.0 and Oximeter 2.0 | ||
| + | # Prepare glycerol stocks from the overnight cultures of the transformed cells (for storage at -20 °C). | ||
| + | # Inoculate 5 ml LB (lysogeny broth) medium (with no antibiotic) from glycerol stocks of both oximeters to prepare two overnight cultures | ||
| + | # Use 5 ml LB culture from previous step to inoculate 200 ml of LB media (containing chloramphenicol) and incubate both YFP and YFP.LVA shake flasks at 30°C in an orbital shaker (200 rpm) for 3-4 hours to an OD600 1-3. | ||
| + | # Use 20 ml of over-day LB culture to inoculate 200 ml of defined media, (containing chloramphenicol) and incubate at 30 °C for 18-22 hours until OD600 is between 1.5 - 2.0 | ||
| + | # Transfer the 20 ml culture from shake flask to two bioreactors and add 900 ml of defined medium, followed by 180 μl of polypropylene glycol | ||
| + | # Start fermentation immediately with optimal growth conditions according to the bioreactor fermentation protocol | ||
| + | # Check initial DOT (dissolved oxygen concentration) and maintain the DOT level AT 100% for the first two hours and then collect the first cell sample (2 ml) | ||
| + | # Set DOT level to decrease by 25% every 2 hours for the next 8 hours until DOT reaches 0% (to induce hypoxia) and maintain this DOT | ||
| + | # Collect 2 ml samples from both bioreactors at two hours intervals | ||
| + | # Use 1 ml of the cell sample to take OD (optical density) measurements and use the other 1 ml for centrifugation to pellet the cell and store it at -20 °C | ||
| + | # Resuspend stored cell pellets in 400 μl of distilled water and load 200 μl of each cell sample on a 96 well for a YFP scan using the Tecan Safire2 microplate reader | ||
| + | # Analyse the fluorescence scans to compare the half-life of the fluorescence proteins | ||
</div> | </div> | ||
{{:Team:UCL_London/Template/Footer}} | {{:Team:UCL_London/Template/Footer}} | ||
Revision as of 02:05, 22 September 2011
Contents |
Modifying Oximeter in the lab
Aim: To modify and improve the oximeter device (BBa_K239011) using modified nark promoter (BBa_K239006) submitted by 2009 UCL iGEM team. Changes included replacing the normal GFP (green fluorescent protein) with an enhanced YFP (yellow fluorescent protein), which has an LVA tag added to its C terminal.
Parts used
Part Name |
Parts ID |
Parts Description |
Length (bp) |
Plasmid Backbone |
mNARK promoter |
promoter sensitive to hypoxia |
89 |
on pUC57 (2710 bp) - ampicillin resistance |
|
RBS |
ribosome binding site |
12 |
on pSB1A2 (2079 bp) - ampicillin resistance |
|
YFP.LVA |
enhanced yellow fluorescent protein with LVA tag |
759 |
on pSB1A2 (2079 bp) |
|
Terminator |
double terminator (B0010-B0012) |
129 |
on pSB1A2 (2079 bp) |
|
n/a |
standard plasmid backbone (kanamycin resistance) |
2204 |
n/a |
|
n/a |
standard plasmid backbone (tetracycline resistance) |
2463 |
n/a |
|
n/a |
standard plasmid backbone (chloramphenicol resistance) |
2070 |
n/a |
Procedure for preparing BioBrick
- Carried out digestion of the mNARK promoter and YFP.LVA protein on their relevant plasmid backbones with EcoRI and SpeI. Also digested RBS and Terminator on their respective plasmid backbones with XbaI and PstI
- Carried out digestion of linearised pSB1K3, pSB1T3, pSB1C3 plasmid backbones with EcoRI and PstI
- Performed gel electrophoresis with a sample from all the digestion mixtures to confirm the parts
- Carried out ligation of the mNARK, RBS and pSB1K3 plasmid. Also ligated YFP.LVA, terminator and pSB1T3.
- Transformed separate stocks of competent TOP10 E. coli cells with the ligation mixtures and grew them overnight on antibiotic plates and then selective LB medium.
- Mini-prepped the overnight cultures to extract the plasmids.
- Digested the mNark/RBS/pSB1K3 with EcoRI and SpeI. Also digested YFP.LVA/Terminator/pSB1T3 with XbaI and PstI
- Performed gel electrophoresis with a sample from both digestion mixtures to confirm the composite parts
- Carried out ligation of mNark/RBS, YFP.LVA.TER and pSB1C3
- Transformed a stock of competent TOP10 E. coli cells with the ligation mixture and grew the cells overnight on chloramphenicol plates, followed by overnight culture in selective LB medium
- Set up a glycerol stock from overnight culture (for storage at -80 °C) and mini-prepped the rest to extract plasmid with the final construct into 50 μl sample
- Carried out digestion of mini-prep sample with EcoRI and PstI and performed gel electrophoresis to confirm the length of the final construct on gel
- Sent a sample of the mini-prep for sequencing to confirm the final construct
- Submitted the improved BioBrick (BBa_K676002) to the registry
Parts submitted to Registry
Part Name |
Parts ID |
Parts Description |
Length (bp) |
Plasmid Backbone |
mNARK with YFP.LVA |
Improved oximeter for detecting hypoxia and express YFP |
1014 |
on pSB1C3 (2070 bp) - chloramphenicol resistance |
Characterisation Plan for the new improved Oximeter
Aim: To compare the sensitivity and half-life of fluorescence assays between Oximeter 2.0 (mNark/RBS/YFP.LVA/Terminator on pSB1C3) and Oximeter 1.0 (mNark/RBS/YFP/Terminator on pSB1C3)
- Carry out 3A assemblies to construct the Oximeter 1.0 device with wild type YFP (yellow fluorescent protein)
- Transform two separate stocks of competent TOP10 E. coli cells with the mini-prep DNA for Oximeter 1.0 and Oximeter 2.0
- Prepare glycerol stocks from the overnight cultures of the transformed cells (for storage at -20 °C).
- Inoculate 5 ml LB (lysogeny broth) medium (with no antibiotic) from glycerol stocks of both oximeters to prepare two overnight cultures
- Use 5 ml LB culture from previous step to inoculate 200 ml of LB media (containing chloramphenicol) and incubate both YFP and YFP.LVA shake flasks at 30°C in an orbital shaker (200 rpm) for 3-4 hours to an OD600 1-3.
- Use 20 ml of over-day LB culture to inoculate 200 ml of defined media, (containing chloramphenicol) and incubate at 30 °C for 18-22 hours until OD600 is between 1.5 - 2.0
- Transfer the 20 ml culture from shake flask to two bioreactors and add 900 ml of defined medium, followed by 180 μl of polypropylene glycol
- Start fermentation immediately with optimal growth conditions according to the bioreactor fermentation protocol
- Check initial DOT (dissolved oxygen concentration) and maintain the DOT level AT 100% for the first two hours and then collect the first cell sample (2 ml)
- Set DOT level to decrease by 25% every 2 hours for the next 8 hours until DOT reaches 0% (to induce hypoxia) and maintain this DOT
- Collect 2 ml samples from both bioreactors at two hours intervals
- Use 1 ml of the cell sample to take OD (optical density) measurements and use the other 1 ml for centrifugation to pellet the cell and store it at -20 °C
- Resuspend stored cell pellets in 400 μl of distilled water and load 200 μl of each cell sample on a 96 well for a YFP scan using the Tecan Safire2 microplate reader
- Analyse the fluorescence scans to compare the half-life of the fluorescence proteins
 "
"