Team:UCL London/Research/Stresslights/Theory
From 2011.igem.org
Contents |
Shear Stress: Shearometer
A. Spy Promoter
A common drawback in high-density bioreactor runs is the shear stress imparted by the impeller blades to the cultivating Escherichia coli cells. This inflicts damage to the bacterial cell wall, exposes its cell membrane and thus makes the cell osmotically fragile and prone to lysis. However, E. coli cells already have a built in protective mechanism in which they can detect the presence of heterogenous unfolded proteins, from the sheared outer membrane, in the periplasmic space and activate the expression of particular proteins to counter the problem. One of this protein is the Spy (Spheroplast protein Y), which was first discovered exclusively in cells which were being subjected to sphaeroplasting stress [1]. Further investigation has revealed the function of Spy protein to be quite similar to molecular chaperones and it represses protein aggregation from sphaeroplasting by aiding protein folding [2].
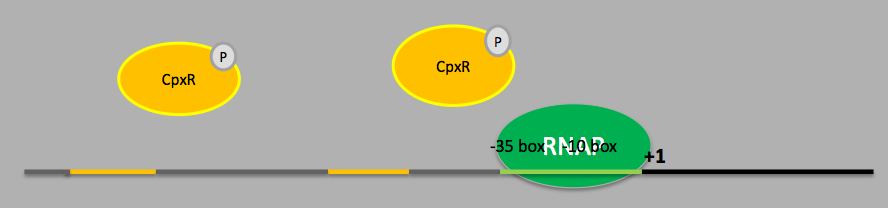
Following this discovery, the Spy transcriptional activation mechanism has been studied thoroughly and it has been indicated that the Spy promoter sequence contains 2 phosphorylated BaeR (Bacterial Adaptive rEsponse Regulator) binding sites, 1 phosphorylated CpxR (Conjugative Plasmid eXpression Regulator) binding site and transcription is initiated under sigma factor 70 (σ70) [3]. For the conjugative plasmid expression pathway, the CpxA-CpxR dimer plays the role of the signal transduction system. The CpxA (a histidine kinase) is the sensor unit which auto-phosphorylates a conserved histidine residue when proteins designated for the outer membrane or secretion (e.g. pili or curli fibres) are mounting in the periplasm. The phosphate from the CpxA histidine is then transferred to a conserved aspartate residue on CpxR, which becomes activated as a DNA binding protein and acts as a transcriptional regulator for various genes [4]. The genes activated are mostly chaperones and proteases involved in the refolding or degradation of accumulated proteins in the periplasm. Further studies carried out by researchers indicated that the level of consensus within the proposed sequence for the CpxR binding site and its orientation seems to play a minor role when determining the strength of induction by CpxR for a particular promoter [5]. However the location of the binding site seems to be of more importance instead.
On the other hand, the bacterial adaptive response pathway was identified later on and it involves a two-component signal transduction system as well. BaeS-R dimer is activated in a similar way as the CpxA-R dimer by stimuli like PapG overexpression, spheroplast formation and indole [3]. However, BaeS-R activation pathway is less well characterised and so far BaeR is known to activate only 4 operons (spy, mdtA, arcD and ycaC) with no known negative regulatory mechanism as yet. Analysis of the upstream region of Spy, mdtA and arcD has led to the identification of a 18 bp long binding sequence for BaeR [6]. Further experiments on the Spy promoter have also confirmed the promoter sequence TCTNCANAA as the BaeR-box [7]. Currently more transcriptional activators for the Spy locus like RcsB (Regulator of colanic acid Capsule Synthesis) are still being identified [8].
A. DegP Promoter
The DegP protein expressed in E. coli is a periplasmic serine protease necessary for survival at very high temperatures. The main role of this protein is to degrade abnormal and excess proteins present in the periplasmic space [9]. Additionally, DegP has also been demonstrated to possess chaperone activity, which is retained even in proteolytically inactive mutant isoforms [10]. The DegP promoter sequence has been characterised and is known to include 2 binding sites for phosphorylated CpxR (Conjugative Plasmid eXpression) and transcription from this promoter is therefore activated in a very similar way to that of the Spy promoter [11]. However, the DegP promoter binds to sigma factor 24 (σ24) factor instead of sigma factor 70 (as in the case of the Spy promoter) and therefore expression from this promoter is dependent on the growth phase of the cell and cannot be activated by the presence of an excess of phosphorylated CpxR alone [12]. Hence the degP promoter activity is only displayed during the stationary phase of the cell.
Hypoxic Stress: Oximeter
A. Nark & mNark Promoter
Another common issue faced in industrial grade bioreactor fermentations is the presence of dead spots inside the fermenter vessel where oxygen is not reaching in sufficient amount. Even though electronic probes can determine the amount of dissolved oxygen in the culture broth accurately, in large volumes the presence of hypoxic regions is inevitable and cannot be detected always by oxygen probes alone. Experiments carried out at the University of California have showed that NarK protein expression in E. coli is upregulated 100-fold under anaerobic conditions and a further 8-fold in the presence of nitrate [13]. Therefore the Nark promoter can essentially be used as a sensory tool for detecting hypoxic regions inside a bioreactor.
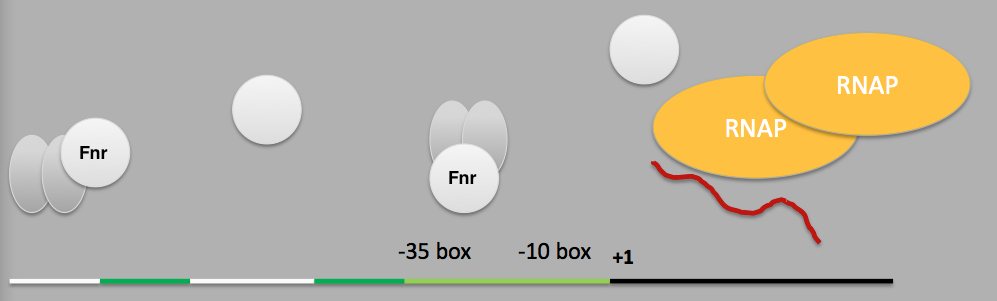
Since then the nark promoter sequence has been studied and is now known to contain 2 Fnr (fumarate and nitrate reductase), 1 Fis, 4 NarL and 1 IHF binding sites that initiate transcription from the promoter by sigma factor 70. The chief protein in this sensory pathway is Fnr which is always present within the cell in a fairly large and constant quantity. In the presence of oxygen, the [4Fe-4S]2+ cluster of Fnr monomer is oxidised to a [2Fe-2S]2+ cluster and the protein therefore remains in their inactive monomer state [14]. But in the absence of oxygen, the [4Fe-4S]2+ cluster is retained and the Fnr protein dimerises and can then bind to DNA sequences and act as a transcriptional activator for various genes.
The optimal and conserved Fnr binding sequence has been determined as TTGATNNNNATCAA [15]. But the distal Fnr binding site in the E. coli NarK promoter is slightly different from the consensus region and therefore has a lower affinity for Fnr. For this reason, a modified NarK promoter sequence was designed in which the distal binding site was re-written to the optimal sequence and also some of the excess nucleotides were removed. As a result, the final mNark promoter sequence is shorter and has a better affinity for activated Fnr and therefore, has a stronger activity under anaerobic condition.
Table: Summary of the binding site for the different regulatory proteins for the three natural promoter sequences (Information collected from EcoCyc database).
LVA Tag
Fluorescent proteins (e.g. GFP, YFP, etc.) have been widely used since their discovery in 1961 as reporter proteins to monitor various cellular processes [16]. However there is one major flaw of these proteins, which is that they are quite stable and have a long half-life. As a result, the fluorescent readings measured from a cell sample may not be quite accurate due to the presence of residual proteins which was expressed at a previous time point for sample collection. Therefore, this property renders them unsuitable for monitoring and quantifying rapid changes in gene expression. However researchers have come up with a solution by demonstrating that the addition of a protease tag (amino acid sequence) to these proteins targets them for degradation by proteases and hence reduces their half-life. Among all the protease tags identified so far, the LVA tag has proven to be the most efficient in making GFP (the most commonly used variety of fluorescent protein) unstable [17]. This so called LVA tag consists of a short peptide sequence (AANDENYALVA) and is added to the C-terminal end of a GFP, which is then targeted for degradation by tail-specific proteases like Tsp protease in the periplasmic space and ClpXP and ClpAP in the cytoplasm of E. coli [17]. Faster degradation using this method would therefore ensure more sensitive and precise fluorescence assays for our sensory devices.In our lab we decided to use the yellow mutant of the natural GFP called the YFP (yellow fluorescent protein). In a research conducted by Jeffrey et al, the half-life of YFP determined in-vivo was 12.8 hours which was later effectively reduced to 5.6 hours after the addition of the LVA tag [18]. The graph below illustrates the fluorescent comparison between YFP and YFP -LVA and it indicated that that the LVA tag reduces the half-life by a factor of 2.28. The fluorescence is still visible for YFP after 30 hours whereas it is 0 for YFP -LVA.
 "
"