Team:UQ-Australia/Data
From 2011.igem.org
(Difference between revisions)
(Created page with "{| style="color:#7B68EE;background:#E6E6FA" cellpadding="5" cellspacing="2" border="2" bordercolor="#4B0082" width="100%" align="center" !align="center"|[[Team:UQ-Australia|Home]...") |
|||
| (27 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {| style=" | + | {{:Team:UQ-Australia/Template:Header}} |
| - | + | {|style="width:100%;" border="0" cellpadding="10" cellspacing="0" | |
| - | + | |- valign="top" | |
| - | + | |rowspan="2"|A brief summary of some of our laboratory findings is included below. For more information, see the [[Team:UQ-Australia/Project|Project]] page. From these gels, you can see the amplification of our genes of interest, which we achieved with primers that added the standard BioBrick prefix and suffix to each end of the genes. As a result, these constructs are all ready for ligation and the construction of our oscillator modules! | |
| - | + | ![[File:UQ-Australia_logo_2011.png|125x125px|link=https://2011.igem.org/Team:UQ-Australia]] | |
| - | + | |- | |
| - | ! | + | | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| + | |||
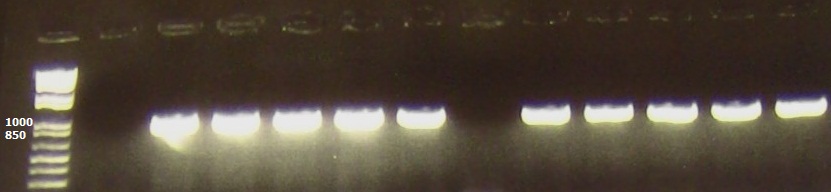
| + | We conducted a heat cycle PCR to determine the optimum conditions for amplifying our araC gene. The lanes here are loaded as follows: | ||
| + | |||
| + | 1: 1kb+ ladder | ||
| + | |||
| + | 2: Negative control (H2O) | ||
| + | |||
| + | 3-7: pBAD33 plasmid at temperatures 50C, 55C, 58C, 60C and 65C. | ||
| + | |||
| + | 8: Negative control (H2O) | ||
| + | |||
| + | 9-13: pBAD33 plasmid at temperatures 50C, 55C, 58C, 60C and 65C. | ||
| + | |||
| + | The expected 900bp band for araC is seen at each temperature | ||
| + | |||
| + | [[File:AraC.jpg| 500x150px]] | ||
| + | |||
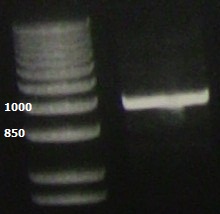
| + | PCR of the lacI gene showing the expected ~1kb band. | ||
| + | |||
| + | [[File:Laci.jpg]] | ||
| + | |||
| + | PCR of glnG (lanes 2 and 3), glnAp2 (lanes 5 and 6), and the promoter Plac/ara (lanes 2-4 of the second gel). Only glnG is the expected size (1.6kb), while the other two significantly smaller parts (200-300bp) have not been amplified. | ||
| + | |||
| + | [[File:Glngplac.jpg | 600x400px]] | ||
Latest revision as of 00:39, 6 October 2011
 "
"