Team:Paris Bettencourt/Designs
From 2011.igem.org
| (116 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Paris_Bettencourt/tpl_test}} | {{:Team:Paris_Bettencourt/tpl_test}} | ||
<html> | <html> | ||
| + | <h1>Designs overview</h1> | ||
| + | <h2>Introduction</h2> | ||
| - | <p> | + | <table> |
| + | <tr> | ||
| + | <td> | ||
| + | <p><b>Our goal for this summer is to characterize the travel of various molecules through nanotubes. In order to do so, we developed several designs. In this page, we explain the principle of the different designs we have built.</b></p> | ||
| - | < | + | <p>In the original paper <a href="https://2011.igem.org/Team:Paris_Bettencourt/Designs#references">[1]</a>, the authors directly observed the passage of molecules through the nanotubes. The idea behind our approach is to use <em>synthetic biology methods</em> to improve the sensitivity and the resolution of these experiments. Relying on <em>signal amplification</em> and natural, as well as artificial, <em>bistable switches</em>, we want to detect, with a resolution at the molecular level, the passage from <b>one emitter cell</b> to <b>the receiver</b>.</p> |
| - | <a href="https://2011.igem.org/ | + | </td> |
| + | <td style="width:200px;"><center><img src="https://static.igem.org/mediawiki/2011/8/86/Logo_projet.png" alt="our logo" width="150px"></center></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <h2>Main questions</h2> | ||
| - | <p>The | + | <p>The nanotube discovery is very recent. Lots of questions remain unsolved concerning the mechanism behind the formation of the tubes, the <em>efficiency of the transfer</em>, the extent of the process. The existence of the tubes themselves remains a subject of controversy within the scientific community.</p> |
| - | <p>We | + | <p>We aim to bring additional proof of their existence and better characterize the extent of this phenomenon. Here is the list of the questions we have been asking ourselves, and that our designs aim to answer:<p> |
| + | |||
| + | <table> | ||
| + | <tr> | ||
| + | <td style="width:200px; text-align:center"><img style="width:150px" src="https://static.igem.org/mediawiki/2011/2/2b/Question_mark_button.png"> | ||
| + | </td> | ||
| + | <td> | ||
<ul> | <ul> | ||
| - | <li>< | + | <li>What is the <em>nature</em> of the process? (passive or active transport)</li> |
| - | <li> | + | <li>What kinds of molecules can pass through the nanotubes?</li> |
| + | <li>Does the communication happen only between <i>B. subtilis</i>? Or can it also exist between different species?</li> | ||
| + | <li>If a communication can be established with Gram negative bacteria, is the communication happening through the periplasm or the cytoplasm?</li> | ||
</ul> | </ul> | ||
| - | < | + | </td> |
| + | </tr> | ||
| + | </table> | ||
| + | <p>There is also the question of the formation of the tubes. However, this specific problem would require a lot more than a summer and a team of undergraduates to be fully investigated. We therefore choose not to focus too much on it, even though we have some <a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion">ideas</a>. | ||
| - | < | + | <h2>Specification of our designs</h2> |
| - | |||
| - | < | + | <p>We started designing the project with these previously stated questions in mind. Using several molecules of <em>different sizes and nature</em>, from a T7 RNA polymerase to ComS, we aim to test the <em>nature, size and number of the molecules we want to diffuse through the nanotubes</em>.</p> |
| - | + | ||
| - | <p> | + | <p>We decided that our constructs should follow the global idea summed up in the following scheme:</p> |
| - | <p> | + | <center><a href="https://2011.igem.org/File:Step1_principle.jpg"><img src="https://static.igem.org/mediawiki/2011/e/e6/Schema-presentation.png"></a></center></br> |
| + | <p><center><b>General plan for the experiment design to characterize the nanotubes</center></b></p> | ||
| - | <p> | + | <p>As explained in our <a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling">modeling section</a>, diffusion seems to be too quick comparatively to genetic networks response time to be accurately measured. We therefore focused on testing the <em>nature</em>, the <em>number</em> and the <em>size</em> of the molecules we try to pass trough the nanotubes.</p> |
| - | < | + | <p>In <i>B.subtilis</i>, nanotubes are reported allow transfer to the cytoplasm. We had to find molecules that can be expressed in the first cell and then trigger a genetic circuit in the second cell. The image below illustrates the idea:</p> |
| - | < | + | <a href="https://2011.igem.org/File:cytoplasm_cytoplasm_communication.jpg"><img src="../wiki/images/4/49/Cytoplasm_cytoplasm_communication.jpg" style="width: 400px; margin-left: 235px;"></a></br> |
| + | <p><center><b>Schematic of the supposed connection between the cells via the nanotubes</b></center></p> | ||
| - | |||
| - | < | + | <h2>Genetic devices we tested</h2> |
| - | + | ||
| - | <p> | + | <p>Using these specifications, we designed several sensitive genetic circuits that can be trigerred by molecules of various sizes. The molecules chosen cover 1 orders of magnitude of radius.</p> |
| + | |||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/d/d7/Size_chart.png" style="width: 900px;"> | ||
| + | </center> | ||
| + | |||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | |||
| + | <table> | ||
| + | |||
| + | <tr> | ||
| + | <th style="width:200px;"></th> | ||
| + | <th style="text-align:center;"><b>Description</b></th> | ||
| + | <th style="width:100px;text-align:center;"><b>Type of switch</b></th> | ||
| + | <th style="width:100px;text-align:center;"><b>B. subilis/<br/>E. coli<br/>compatibility</b></th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="text-align:center;"><a href="https://2011.igem.org/Team:Paris_Bettencourt/T7_diffusion"><img style="width:150px; margin-top:20px;" src="https://static.igem.org/mediawiki/2011/e/e4/T7_button.png"></a> | ||
| + | </td> | ||
| + | <td><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/T7_diffusion">T7 polymerase diffusion</a></em> The T7 RNA polymerase is the RNA polymerase of the T7 phage. This is a big molecule, that recognizes a very specific promoter orthogonal to <i>B.subtilis</i>. | ||
| + | </td> | ||
| + | <td style="text-align:center;">Irreversible<br/>amplification</td> | ||
| + | <td style="text-align:center;">Yes</td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="text-align:center"><a href="https://2011.igem.org/Team:Paris_Bettencourt/tRNA_diffusion"><img style="width:150px; margin-top:20px;" src="https://static.igem.org/mediawiki/2011/5/53/TRNAamber-button.png"></a> | ||
| + | </td> | ||
| + | <td><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/tRNA_diffusion">Amber suppressor tRNA diffusion</a></em> The principle of this design is to produce in one cell an amber supressor tRNA that will diffuse through the nanotubes. The receptor cell holds the gene for T7 with amber stop codons that cannot be translated into a functional protein as long as the tRNA amber suppressor is not present in the cell. Once expressed, the functional amber T7 RNA polymerase will trigger the T7 amplification system. | ||
| + | </td> | ||
| + | <td style="text-align:center;">Irreversible<br/>amplification</td> | ||
| + | <td style="text-align:center;">Yes</td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="text-align:center"><a href="https://2011.igem.org/Team:Paris_Bettencourt/SinOp"><img style="width:150px; margin-top:20px;" src="https://static.igem.org/mediawiki/2011/a/aa/SinOp-button.png"></a> | ||
| + | </td> | ||
| + | <td><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/SinOp">KinA diffusion</em></a> The Sin operon system controls the sporulation switch of <i>B. subtilis</i>. Making it diffuse from a non sporulating strain to a receiver cell makes the receiver cell to sporulate. We also use fluorescent sporulation reporters to monitor the experiment properly. | ||
| + | </td> | ||
| + | <td style="text-align:center;">Switch</td> | ||
| + | <td style="text-align:center;">Mono-<br/>directional</td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="text-align:center"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/Lambda_switch"><img style="width:150px; margin-top:20px;" src="https://static.igem.org/mediawiki/2011/9/94/Lambda_switch-button.png"></a> | ||
| + | </td> | ||
| + | <td><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/Lambda_switch">Lambda switch design</em></a> The CI is a regulation protein from the lambda phage switch. Make it diffuse would change the state of the toogle switch from the push-on push-off system, from the <a href="https://2007.igem.org/Peking">PKU 2007</a>. | ||
| + | </td> | ||
| + | <td style="text-align:center;">Switch</td> | ||
| + | <td style="text-align:center;">Mono-<br/>directional</td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | <tr> | ||
| + | <td style="width:200px; text-align:center"><a href="https://2011.igem.org/Team:Paris_Bettencourt/GFPLac_diffusion"><img style="width:150px" src="https://static.igem.org/mediawiki/2011/d/d0/YFP_concentration_button.png"></a> | ||
| + | </td> | ||
| + | <td><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/GFPLac_diffusion">YFP-TetR/TetO array experiment</a></em> This experiment is an improvement of the GFP diffusion experiment of the original paper. To observe significant fluorescence in the neighboring cells, lots of molecules have to pass through the tubes. Using the affinity of the TetR for the TetO array, we want to concentrate in one spot the YFP molecules to better monitor this diffusion. | ||
| + | <td style="text-align:center;">Concentrator</td> | ||
| + | <td style="text-align:center;">Yes</td> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | <tr> | ||
| + | <td style="width:200px; text-align:center"><a href="https://2011.igem.org/Team:Paris_Bettencourt/ComS_diffusion"><img style="width:150px; margin-top:20px;" src="https://static.igem.org/mediawiki/2011/2/21/ComS-button.png"></a> | ||
| + | </td> | ||
| + | <td><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/ComS_diffusion">ComS diffusion</a></em> ComS is an inhibitor of the MecA protease in <i>B.subtilis</i>. It plays a key role in the triggering of the competence and sporulation mechanisms. The idea is to trigger the switch of the MeKS system of the receptor cell by diffusing ComS through the nanotubes. | ||
| + | </td> | ||
| + | <td style="text-align:center;">Switch</td> | ||
| + | <td style="text-align:center;">Mono-<br/>directional</td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | |||
| + | <h2>Feedback from the models</h2> | ||
| + | |||
| + | <p>Our <a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling">modeling</a> showed us which designs would theoritically be the easiest to monitor and which one might only lead to inconclusive results.<p> | ||
| + | |||
| + | <p>The feedback from our models showed us that: | ||
<ul> | <ul> | ||
| - | <li> | + | <li><em>Passive diffusion</em> alone can explain the results observed in the Dubey/Ben-Yehuda article, although other processes might contribute significantly</li> |
| - | <li> | + | <li><em>Diffusion times</em> are too short compared to genetic response for us to monitor properly</li> |
| - | <li> | + | <li>All our systems will respond in <em>approximately one hour</em> which fits nicely with the timescale of the GFP diffusion observed in the Dubey/Ben-Yehuda article</li> |
| - | <li> | + | <li>We know roughly <em>how many molecules will be required to activate</em> each of our system (500 for ComS diffusion, 10 for T7 RNA polymerase, etc.)</li> |
| - | <li> | + | <li>Since we change experimental conditions for the Coms diffusion system and the Sin Operon system, we know those <em>two systems might not work as expected</em></li> |
</ul> | </ul> | ||
| - | < | + | </p> |
| + | <p>Using all these modeling results, we were able to <em>design properly our experiments</em> and have some expectations as to the results.</p> | ||
| - | <h2> | + | <h2>Others designs not built</h2> |
| - | <p>We | + | <p>We had not the manpower to try all the ideas we had. We designed some systems in order to investigate the <i>E. coli / B. subtilis</i> connection nature but had no time to investigate this question further. Nonetheless, we present some of our design ideas for solving this problem in this section.</p> |
| - | < | + | <p><i>B.subtilis</i> is Gram- so the nanotubes seem to create a link from the cytoplasm of the first cell to the cytoplasm of the second cell. In the case of the Gram negative bacteria, like<i> E.coli</i>, we wonder how the nanotubes connect the cells. Do they create a link with the periplasm or with the cytoplasm? To test the two hypotheses, we tried to create a design for each of the two possibilities. If one work and the other does not, the answer to this question will be known.</p> |
| - | < | + | <h3>Design for <i>B.subtilis / E.coli</i> if the connection happens with the cytoplasm</h3> |
| - | < | + | |
| - | + | <p>If the connection between <i>E.coli</i> and <i>B.subtilis</i> is happening through the cytoplasm, the type of connection could be summed up by the following schematic.</p> | |
| + | |||
| + | <img src="../wiki/images/3/3a/Cytoplasm_connection_with_coli.jpg" style="width: 500px; margin-left: 185px;"> | ||
| + | <p><center><b>Schematics of the communication happening directly to the cytoplasm</b></center></p> | ||
| + | |||
| + | <p>In this case, be can re-use most of the designs of the first section (see the compatibility column). We also have proposed this additional design:</p> | ||
| - | |||
| - | |||
<ul> | <ul> | ||
| - | <li> | + | <li><a href="https://2011.igem.org/Team:Paris_Bettencourt/Xis_diffusion">Xis protein diffusion</a> Xis is a small partner of an excisase. The latter will excise a stop codon on the DNA strand that is preventing the expression of the GFP. We had no time to build this design, but the idea is summed up <a href="https://2011.igem.org/Team:Paris_Bettencourt/Xis_diffusion">here</a>.</li> |
| - | + | ||
</ul> | </ul> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | <p> | + | <h3>Design for <i>B.subtilis / E.coli</i> if the connection happens with the periplasm</h3> |
| + | |||
| + | <p>In case the communication happens with the periplasm, we had to think about molecules that can be then transported into the cytoplasm.</p> | ||
| + | |||
| + | <a href="https://2011.igem.org/File:Periplasm_conection.jpg"><img src="https://static.igem.org/mediawiki/2011/4/40/Periplasm_conection.jpg" style="width: 500px; margin-left: 185px;"></a></br> | ||
| + | <p><center><b>Schematic of the communication happening through the periplasm.</b></center></p> | ||
| + | |||
| + | <p>We had to design more sophisticated approaches. The ideas are the following:</p> | ||
<ul> | <ul> | ||
| - | <li> | + | <li><a href="https://2011.igem.org/Team:Paris_Bettencourt/MPB_diffusion">MBP diffusion:</a> we need a CRP+, MBP- <i>E.coli</i> mutant. We produce the MBP protein in <i>B.subtilis</i> and make it diffuse through the nanotubes. As long as the MBP has not reached the periplasm of <i>E.coli</i>, the cell cannot digest the maltose in the medium. The indirect induction of MalR by MBP triggers the expression of the GFP reporter.</li> |
| - | + | ||
</ul> | </ul> | ||
| - | < | + | <br> |
| + | |||
| + | |||
| - | |||
<br> | <br> | ||
| - | <h1>Designs for a Master-Slave system</h1> | + | <!--<h1>Designs for a Master-Slave system</h1> |
| - | <p>The principle of Step 2 is to build a Master-Slave system, where the Master controls the state of the Slave cell in a | + | <p>The principle of Step 2 is to build a Master-Slave system, where the Master controls the state of the Slave cell in a mono-directional exchange.</p> |
<a href="https://2011.igem.org/File:Master_Slave_system_principle.jpeg"><img src="../wiki/images/5/5a/Master_Slave_system_principle.jpeg" style="width: 600px; margin-left: 185px;"></a></br> | <a href="https://2011.igem.org/File:Master_Slave_system_principle.jpeg"><img src="../wiki/images/5/5a/Master_Slave_system_principle.jpeg" style="width: 600px; margin-left: 185px;"></a></br> | ||
| - | + | <p><center><b>Specification of the master slave design</b></center></p> | |
<p>Such a design implies the reversibility of all the sub-systems, the activators and the amplifiers in particular.</p> | <p>Such a design implies the reversibility of all the sub-systems, the activators and the amplifiers in particular.</p> | ||
| Line 93: | Line 201: | ||
<p>Several potential designs are summed-up below:</p> | <p>Several potential designs are summed-up below:</p> | ||
<ul> | <ul> | ||
| - | <li> | + | <li><a href="https://2011.igem.org/Team:Paris_Bettencourt/ComS_diffusion">The ComS system:</a> The characterization step of the MeKS system turn to be reversible. It can be considered as well as a Master/Slave system.</li> |
| - | + | </ul>--> | |
| - | </ul> | + | |
<br> | <br> | ||
| - | < | + | <div id="citation_box"> |
| - | + | <p id="references">References</p> | |
| - | <p> | + | <ol> |
| - | < | + | <li><i>Intercellular Nanotubes Mediate Bacterial Communication</i>, Dubey and Ben-Yehuda, Cell, available <a href="http://bms.ucsf.edu/sites/ucsf-bms.ixm.ca/files/marjordan_06022011.pdf">here</a></li> |
| - | + | </ol> | |
| - | + | </div> | |
| - | + | <br> | |
| - | <a href=" | + | |
| - | + | ||
| - | + | ||
| - | < | + | |
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
| Line 147: | Line 250: | ||
</li> | </li> | ||
| - | <li id="t-permalink"><a href="/wiki/index.php? | + | <li id="t-permalink"><a href="/wiki/index.php? |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 01:51, 29 October 2011

Designs overview
Introduction
|
Our goal for this summer is to characterize the travel of various molecules through nanotubes. In order to do so, we developed several designs. In this page, we explain the principle of the different designs we have built. In the original paper [1], the authors directly observed the passage of molecules through the nanotubes. The idea behind our approach is to use synthetic biology methods to improve the sensitivity and the resolution of these experiments. Relying on signal amplification and natural, as well as artificial, bistable switches, we want to detect, with a resolution at the molecular level, the passage from one emitter cell to the receiver. |
 |
Main questions
The nanotube discovery is very recent. Lots of questions remain unsolved concerning the mechanism behind the formation of the tubes, the efficiency of the transfer, the extent of the process. The existence of the tubes themselves remains a subject of controversy within the scientific community.
We aim to bring additional proof of their existence and better characterize the extent of this phenomenon. Here is the list of the questions we have been asking ourselves, and that our designs aim to answer:

|
|
There is also the question of the formation of the tubes. However, this specific problem would require a lot more than a summer and a team of undergraduates to be fully investigated. We therefore choose not to focus too much on it, even though we have some ideas.
Specification of our designs
We started designing the project with these previously stated questions in mind. Using several molecules of different sizes and nature, from a T7 RNA polymerase to ComS, we aim to test the nature, size and number of the molecules we want to diffuse through the nanotubes.
We decided that our constructs should follow the global idea summed up in the following scheme:

As explained in our modeling section, diffusion seems to be too quick comparatively to genetic networks response time to be accurately measured. We therefore focused on testing the nature, the number and the size of the molecules we try to pass trough the nanotubes.
In B.subtilis, nanotubes are reported allow transfer to the cytoplasm. We had to find molecules that can be expressed in the first cell and then trigger a genetic circuit in the second cell. The image below illustrates the idea:
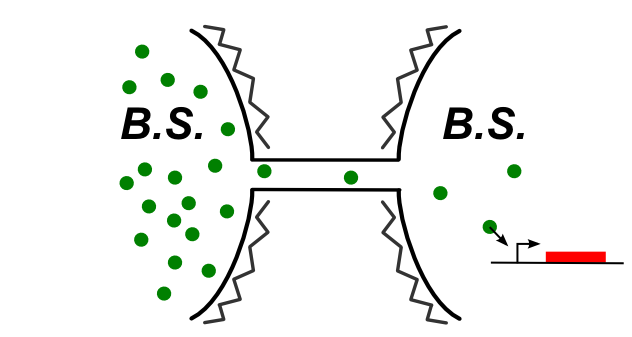
Genetic devices we tested
Using these specifications, we designed several sensitive genetic circuits that can be trigerred by molecules of various sizes. The molecules chosen cover 1 orders of magnitude of radius.

| Description | Type of switch | B. subilis/ E. coli compatibility |
|
|---|---|---|---|

|
T7 polymerase diffusion The T7 RNA polymerase is the RNA polymerase of the T7 phage. This is a big molecule, that recognizes a very specific promoter orthogonal to B.subtilis. | Irreversible amplification |
Yes |

|
Amber suppressor tRNA diffusion The principle of this design is to produce in one cell an amber supressor tRNA that will diffuse through the nanotubes. The receptor cell holds the gene for T7 with amber stop codons that cannot be translated into a functional protein as long as the tRNA amber suppressor is not present in the cell. Once expressed, the functional amber T7 RNA polymerase will trigger the T7 amplification system. | Irreversible amplification |
Yes |

|
KinA diffusion The Sin operon system controls the sporulation switch of B. subtilis. Making it diffuse from a non sporulating strain to a receiver cell makes the receiver cell to sporulate. We also use fluorescent sporulation reporters to monitor the experiment properly. | Switch | Mono- directional |

|
Lambda switch design The CI is a regulation protein from the lambda phage switch. Make it diffuse would change the state of the toogle switch from the push-on push-off system, from the PKU 2007. | Switch | Mono- directional |

|
YFP-TetR/TetO array experiment This experiment is an improvement of the GFP diffusion experiment of the original paper. To observe significant fluorescence in the neighboring cells, lots of molecules have to pass through the tubes. Using the affinity of the TetR for the TetO array, we want to concentrate in one spot the YFP molecules to better monitor this diffusion. | Concentrator | Yes |

|
ComS diffusion ComS is an inhibitor of the MecA protease in B.subtilis. It plays a key role in the triggering of the competence and sporulation mechanisms. The idea is to trigger the switch of the MeKS system of the receptor cell by diffusing ComS through the nanotubes. | Switch | Mono- directional |
Feedback from the models
Our modeling showed us which designs would theoritically be the easiest to monitor and which one might only lead to inconclusive results.
The feedback from our models showed us that:
- Passive diffusion alone can explain the results observed in the Dubey/Ben-Yehuda article, although other processes might contribute significantly
- Diffusion times are too short compared to genetic response for us to monitor properly
- All our systems will respond in approximately one hour which fits nicely with the timescale of the GFP diffusion observed in the Dubey/Ben-Yehuda article
- We know roughly how many molecules will be required to activate each of our system (500 for ComS diffusion, 10 for T7 RNA polymerase, etc.)
- Since we change experimental conditions for the Coms diffusion system and the Sin Operon system, we know those two systems might not work as expected
Using all these modeling results, we were able to design properly our experiments and have some expectations as to the results.
Others designs not built
We had not the manpower to try all the ideas we had. We designed some systems in order to investigate the E. coli / B. subtilis connection nature but had no time to investigate this question further. Nonetheless, we present some of our design ideas for solving this problem in this section.
B.subtilis is Gram- so the nanotubes seem to create a link from the cytoplasm of the first cell to the cytoplasm of the second cell. In the case of the Gram negative bacteria, like E.coli, we wonder how the nanotubes connect the cells. Do they create a link with the periplasm or with the cytoplasm? To test the two hypotheses, we tried to create a design for each of the two possibilities. If one work and the other does not, the answer to this question will be known.
Design for B.subtilis / E.coli if the connection happens with the cytoplasm
If the connection between E.coli and B.subtilis is happening through the cytoplasm, the type of connection could be summed up by the following schematic.
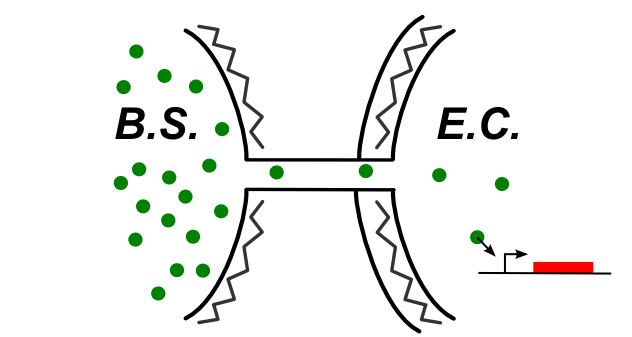
In this case, be can re-use most of the designs of the first section (see the compatibility column). We also have proposed this additional design:
- Xis protein diffusion Xis is a small partner of an excisase. The latter will excise a stop codon on the DNA strand that is preventing the expression of the GFP. We had no time to build this design, but the idea is summed up here.
Design for B.subtilis / E.coli if the connection happens with the periplasm
In case the communication happens with the periplasm, we had to think about molecules that can be then transported into the cytoplasm.

We had to design more sophisticated approaches. The ideas are the following:
- MBP diffusion: we need a CRP+, MBP- E.coli mutant. We produce the MBP protein in B.subtilis and make it diffuse through the nanotubes. As long as the MBP has not reached the periplasm of E.coli, the cell cannot digest the maltose in the medium. The indirect induction of MalR by MBP triggers the expression of the GFP reporter.
References
- Intercellular Nanotubes Mediate Bacterial Communication, Dubey and Ben-Yehuda, Cell, available here
 "
"
