green light receptor
Testdigest with PvuI
Investigators:Julia
digested were Minipreps of Ligationreaction with CcaS.
The Enzym PvuI should cut into two fragments, one of 1666bp, the other 582 bp and vector at 2kb.
Protocol:
for Mastermix of 7 reactions:
30µl H2O
7µl NEB buffer 3
7µl BSA 10x
2µl PvuI
give 3µl of DNA sample to 7µl Mastermix, digest for one hour.
Add 1µl loading dye buffer(6x)a nd let the gel run.
Gelpicture
blue light receptor
Theoretical Gibson-Assembly
Investigators: Sandra, Sophie
Primer Degin for Blue light sensor (lovTAP) + trp promotor (BBa_K322999) and tetR Gen + tetO promotor (BBa_Q04400).
- LOVtap-Trp_up: gaattcgcggccgcttctagtcacacaggaaagtactatgt
- LOVtap-Trp_dw: tacttttatctaatctggacatctagtatttctcctctttgtcgataccctttttacgtg
- TetR-TetO_up: aaagaggagaaatactagatgtccagattag
- TetR-TetO_dw^: ctgcagcggccgctactag
^we also have another primer for this one, but we are not sure about the length/melting temperature. Might be changed.
red light receptor
Testdigest
Investigators: Jakob
| Name: Jakob
| Date: 19.07.2011
|
| Continue from Experiment :
16.07.2011
Miniprep
|
| Project Name: Red light receptor
|
For one reaction you need: For Mastermix: Number of samples+2extra
| 4μl
| H2O
| 24
|
|
| 1μl
| Buffer, NEB4
| 6
|
|
| 1μl
| BSA (10x)
| 6
|
|
| 0,5 μl
| Enzym 1
| 2
|
|
| 0,5 μl
| Enzym 2
| 2
|
|
| 3 μl
| DNA
| 3
|
|
10 μl total volume
Give 3 μl of DNA in an eppi and add 7μl of the mastermix.
Incubate for about 1h at 37°C.
Add 1 μl Loading dye buffer and load the gel.
Picture of the tesdigest: Expected sizes: L22+term(L21) ≈ 800 bp, L23+term(L21) ≈ 800 bp

L22a+b are correct
- To-do: send for sequencing
Miniprep
| Name:
Jakob
| Date:
19.07.2011
|
| Continue from Experiment: 18.07.2011
Trafo and Grow overnight at 37 °C
|
| Project Name:
Red light receptor
|
Documentation:
| Part name: L22+L21 = ho1 + terminator (BBa_I15008 + B1006)
L23+L21 = PcyA + terminator (BBa_I15009 + B1006)
|
Describe your results and mistakes and measure the DNA concentration with the Nanodrop and note the results.
| L22+L21 a-h=ho1+term
| DNA conc. ng/µl
| 260/280
| 260/230
|
| L22+L21 a
| 78
| 1,8
| 1.3
|
| L22+L23 d
| 31,7
| 2,01
| 2,09
|
| L22+L23 f
| 30,4
| 2,58
| -11
|
| L22+L23 g
| 84,8
| 1,98
| 2,58
|
Nothing grew in b,c,e,h
| L23+L21 a-h=PcyA+term
| DNA conc. ng/µl
| 280/230
| 260/230
|
| L23+L21 a
| 87,1
| 1,95
| 2,62
|
| L23+L21 b
| 101,1
| 1,85
| 2,71
|
| L23+L21 c
| 31,4
| 2,91
| 3,40
|
| L23+L21 d
| 184,1
| 1,5
| 0.98
|
| L23+L21 e
| 98
| 1,94
| 2,72
|
| L23+L21 g
| 49,3
| 2,16
| 3,12
|
| L23+L21 h
| 122,3
| 1,94
| 2,77
|
Nothing grew in f
- Labeled: L22+21 a-h, L23+L21 a-h, strored: pink rack -20 freezer
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
PCR
| Name: Ruediger
| Date: 19.07.11
|
| Continue from Experiment (Date)
PCR 18.07 (Name)
|
| Project Name: GFP Pbd
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
| P18, P19, P20
|
| 2.5µl
| Primer dw
| P28
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
| PCR product of P1,P3,S14 from yesterday
|
| 0.5 µl
| Phusion (add in the end)
|
|
What program do you use?
One set of probes was prepared and then split into 3 tubes each to test them at Annealingtemperatures 44C, 52C and 60C for the first 10 cycles. Then Annealingtemperature 60C for 25 cycles
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
S14+P18+P28
S14+P19+P28
S14+P20+P28
Stored in PCR product box
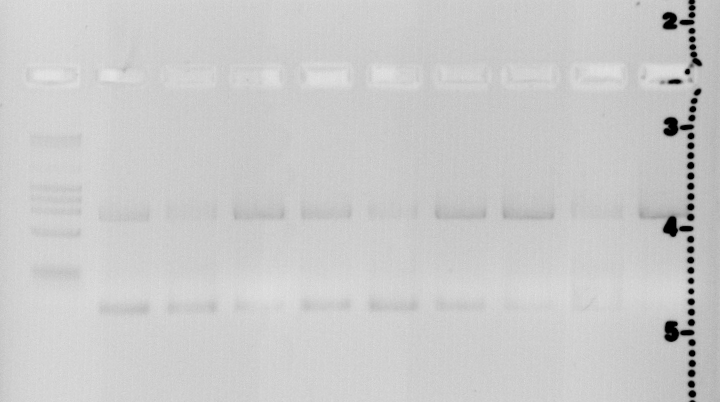
Lane1
Quick Load Marker
Lane2
44C S14+P18+P28
Lane3
44C S14+P19+P28
Lane4
44C S14+P20+P28
Lane5
52C S14+P18+P28
Lane6
52C S14+P19+P28
Lane7
52C S14+P20+P28
Lane8
60 S14+P18+P28
Lane9
60 S14+P19+P28
Lane10
60 S14+P20+P28
Digestion
| Name: Ruediger
| Date: 19.07
|
| Continue from Experiment (Date) 19.07 PCR
(Name) Ruediger
|
| Project Name:
GFP Pbd
|
Procedure
- add H2O (38μl-DNA )
- 5 μl NEB4 buffer (stored at iGEM’s, -20°C)
- 5 μl 10x BSA (used 1:10 diluted sample stored at iGEM’s, -20°C)
- DNA (500 ng)
- 1 μl restriction enzymes (stored at iGEM’s, -20°C)
- heat for 1-2 hours 37°C (6 hours if time)
- heat for 20 minutes 80°C (inactivation of enzymes)
- keep at 4°C if you cannot continue
Vector (ratio 1:3 to insert)
Inserts (500ng)
| Components
| Vector (μl)
| Insert1 (μl)
|
| DNA (500ng)
| 5,6
| 12,5
| 4,7
| 3,5
| 3,8
|
| BSA (100x) (5μl)
|
|
|
|
|
|
| NEB4 Buffer (5μl)
|
|
|
|
|
|
| Enzyme 1 (1μl)
| SpeI
| SpeI
| XbaI
| XbaI
| XbaI
|
| Enzyme 2 (1μl)
| PstI
| PstI
| PstI
| PstI
| PstI
|
| H2O (38 μl- DNA)
| 32,5
| 25,5
| 33,7
| 34,5
| 34,2
|
| In total 50 μl
|
|
|
|
|
|
Measured DNA-concentration with Nanodrop to calculate the volume of DNA to do the digestion:
| Sample
| DNA concentration (μg/μl)
|
| S14+P20+P28 (short:20)
| 132
|
| S14+P19+P28 (short 19)
| 145
|
| S14+P18+P28 (short 18)
| 107
|
| S39
| 90
|
| S43
| 40
|
|
|
|
|
|
|
Documentation:
Why are you doing this experiment? Where are the samples stored? Name of samples, antibiotica resistance, vector used etc.
| Want to ligate GFPpbd (3 different versions P18/19/20) into PR vectors (one with strong Promotor strong RBS, one with middle Promotor, middle RBS)
CM Resistance
Mistake: took 100x BSA instead of 10X
|
 "
"
 Contact
Contact 














